license: mit
pipeline_tag: mask-generation
tags:
- model_hub_mixin
- pytorch_model_hub_mixin
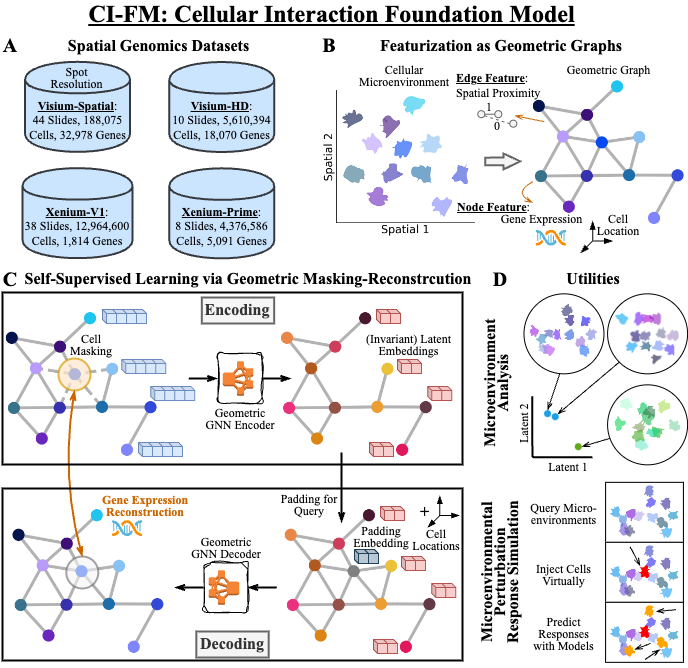
CI-FM: Cellular Interaction Foundation Model
Overview
This is the PyTorch implementation of the CI-FM model -- an AI model that can simulate the cellular activities within a living tissue (AI virtual tissue). The signature functions of CI-FM are:
- Embedding of celllular microenvironments via
embeddings = model.embed(adata)(Figure below panel D top); - Inference of cellular gene expressions at a certain microenvironment via
expressions = model.predict_cells_at_locations(adata, rand_loc)(Figure below panel D bottom).
The detailed usage of the model can be found in the tutorial. Before running the tutorial, please set up an environment following the instruction.
More information about the model can be found in the preprint.
Environment
The environment can be set up via:
pip install torch==2.1.0 --index-url https://download.pytorch.org/whl/cu118
pip install torch-scatter torch-sparse torch-cluster torch-geometric -f https://data.pyg.org/whl/torch-2.1.0+cu118.html
pip install scanpy e3nn
pip install lightning==2.1.0
pip install numpy==1.26.4
pip install transformers
where the compatibility between torch and torch-geometric is not always guaranteed (the first two lines), since these two guys are picky on the platform and the version of other packages. You may trial and error to find the right version of torch and torch-geometric that works for you, e.g., in some machine I once installed via:
pip install torch==2.0.1 --index-url https://download.pytorch.org/whl/cu117
pip install torch-scatter torch-sparse torch-cluster torch-geometric -f https://data.pyg.org/whl/torch-2.0.1+cu117.html
Citation
If you use this code for you research, please cite our paper.
TBD