image
image |
|---|
Overview
This dataset contains Functional Group (FG)-enhanced SMILES designed for molecule representation learning. It consists of approximately 20 million FG-enhanced SMILES collected from a wide range of chemical suppliers and databases, as detailed below:
| Supplier | Number of Compounds | Source |
|---|---|---|
| Targetmol | 22,555 | Targetmol |
| Chemdiv | 1,741,620 | Chemdiv |
| Enamine | 862,698 | Enamine |
| Life Chemical | 347,657 | Life Chemicals |
| Chembridge | 1,405,499 | Chembridge |
| Vitas-M | 1,430,135 | Vitas-M |
| InterBioScreen | 560,564 | InterBioScreen |
| Maybridge | 97,367 | Maybridge |
| Asinex | 601,936 | Asinex |
| Eximed | 61,281 | Eximed |
| Princeton BioMolecular | 1,647,078 | Princeton BioMolecular |
| Otava | 9,203,151 | Otava |
| Alinda Chemical | 733,152 | Alinda Chemical |
| ChEMBL 25 | 1,785,415 | ChEMBL |
| ZINC15 | 4,000,000 | ZINC15 |
| Total | 20,000,000 |
Dataset Details
This dataset contains 36,748 unique tokens (vocabulary size = 36,748), which is an extension from 93 tokens in the corresponding standard SMILES dataset. This expansion helps bridge the gap between SMILES and natural language, which has a large set of vocabulary to sufficiently express the nuances of language.
For more details about the method of generating FG-enhanced SMILES, please refer to our paper, FARM: Functional Group-Aware Representations for Small Molecules, or visit our GitHub repository for implementation details.
Below is a breakdown of the number of token types that represent different chemical elements:
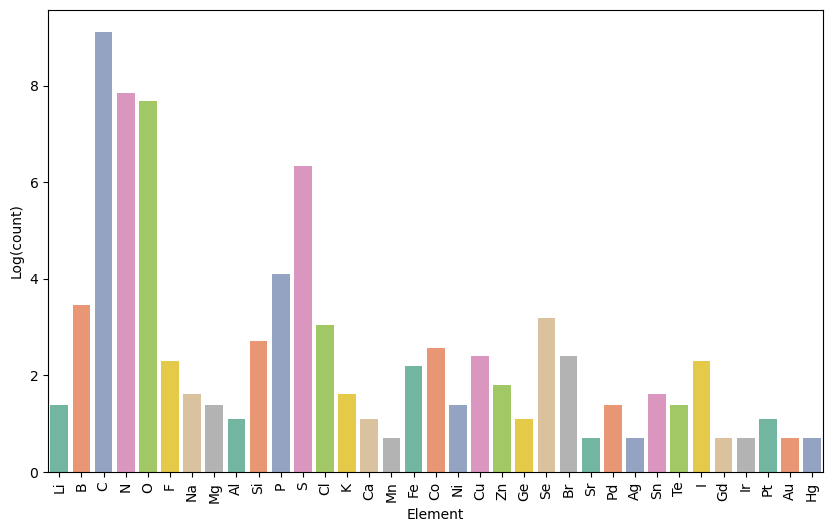 Number of functional groups associated with different chemical elements in the FG-enhanced SMILES dataset. The y-axis represents the natural logarithm (log, base e) of the count.
Number of functional groups associated with different chemical elements in the FG-enhanced SMILES dataset. The y-axis represents the natural logarithm (log, base e) of the count.
Examples of FG-enhanced SMILES
Below are some examples of FG-enhanced SMILES from the dataset. Each SMILES string is augmented with functional group annotations (e.g., C_alkyl, O_ether, c_6, n_tertiary_amine, etc.) to enhance the representation of molecular structures for machine learning models.
C_alkyl O_ether c_6 1 c_6 c_6 c_6 ( - c_5-6 2 n_5-6 c_5-6 3 n_tertiary_amine_5-6 ( C_alkyl c_6 4 c_6 c_6 c_6 c_6 c_6 4 F_fluoro ) c_5-6 ( C_alkyl ) c_5-6 ( - c_5-6 4 c_5-6 c_5-6 c_5-6 5 c_5-6 ( c_5-6 4 ) O_ether_5-6 C_5-6 O_ether_5-6 5 ) c_amide_5-6 ( = O_amide ) n_amide_5-6 3 c_5-6 2 C_alkyl N_tertiary_amine ( C_alkyl ) C_alkyl C_alkyl c_6 2 c_6 c_6 c_6 c_6 c_6 2 ) c_6 c_6 1C_alkyl C_tertiary_carbon ( C_alkyl ) C_tertiary_carbon ( C_ester ( = O_ester ) O_ester C_ester c_5-6 1 c_5-6 n_tertiary_amine_5-6 2 c_5-6 c_5-6 c_5-6 c_5-6 c_5-6 2 n_5-6 1 ) C_tertiary_carbon ( C_alkyl ) N_secondary_amine C_ester ( = O_ester ) O_ester C_ester ( C_alkyl ) ( C_alkyl ) C_alkylC_alkyl C_tertiary_carbon_6 1 C_6 N_tertiary_amine_6 ( c_5 2 n_5 n_5 c_5 ( C_tertiary_carbon_3 3 C_3 C_3 3 ) n_tertiary_amine_5 2 C_alkyl C_alkyl N_secondary_amine C_ketone ( = O_ketone ) c_5 2 c_5 c_5 c_5 s_5 2 ) C_6 C_amide_6 ( = O_amide ) N_amide_6 1 C_alkylC_alkyl O_ether C_alkyl C_alkyl n_tertiary_amine_5 1 c_5 ( C_alkyl c_5 2 c_5 c_5 ( C_alkyl ) n_secondary_amine_5 n_5 2 ) n_5 n_5 c_5 1 N_tertiary_amine_7 1 C_7 C_7 C_7 N_amide_7 ( C_amide ( = O_amide ) C_tertiary_carbon ( C_alkyl ) C_alkyl ) C_7 C_7 1C_alkyl C_alkyl n_amide_6-6 1 c_amide_6-6 ( = O_amide ) n_6-6 c_6-6 ( O_hydroxyl ) c_6-6 2 c_6-6 ( C_ketone ( = O_ketone ) N_secondary_amine N_tertiary_amine_5 3 C_5 C_5 C_5 C_tertiary_carbon_5 3 C_alkyl O_ether C_alkyl ) c_6-6 c_6-6 ( C_tertiary_carbon ( C_alkyl ) C_alkyl ) n_6-6 c_6-6 1 2C_alkyl C_tertiary_carbon_3 1 C_3 C_tertiary_carbon_3 1 C_alkyl n_tertiary_amine_5 1 c_5 ( C_alkyl c_6 2 c_6 c_6 n_6 c_6 c_6 2 ) n_5 n_5 c_5 1 N_tertiary_amine_6 1 C_6 C_6 N_tertiary_amine_6 ( C_alkyl C_amide ( = O_amide ) N_amide ( C_alkyl ) C_alkyl ) C_6 C_6 1
- Downloads last month
- 86