The viewer is disabled because this dataset repo requires arbitrary Python code execution. Please consider
removing the
loading script
and relying on
automated data support
(you can use
convert_to_parquet
from the datasets library). If this is not possible, please
open a discussion
for direct help.
🧠 Dataset Summary
3794 anonymized 3D structural MRI brain scans (T1-weighted MPRAGE NIfTI files) from 2607 individuals included in five publicly available datasets: DLBS, IXI, NKI-RS, OASIS-1, and OASIS-2. Subjects have a mean age of 45 ± 24. 3529 scans come from cognitively normal individuals and 265 scans from individuals with an Alzheimer's disease clinical diagnosis. Scan image dimensions are 113x137x113, 1.5mm^3 resolution, aligned to MNI152 space (see methods).
Scans have been processed and all protected health information (PHI) is excluded. Only the skull-stripped scan, integer age, biological sex, clinical diagnosis, and scan metadata are included. Radiata compiles and processes publicly available neuroimaging datasets to create this open, unified, and harmonized dataset. For more information see https://radiata.ai/public-studies. Example uses include developing foundation-like models or tailored models for brain age prediction and disease classification.
License
The use of the dataset as a whole is licensed under the ODC-By v1.0 license. Individual scans are licensed under study-specific data use agreements:
IXI - CC BY-SA 3.0
DLBS - CC BY-NC 4.0
NKI-RS - Custom DUA
OASIS-1 - Custom DUA
OASIS-2 - Custom DUA
The metadata provide the license for each object.
Sample images
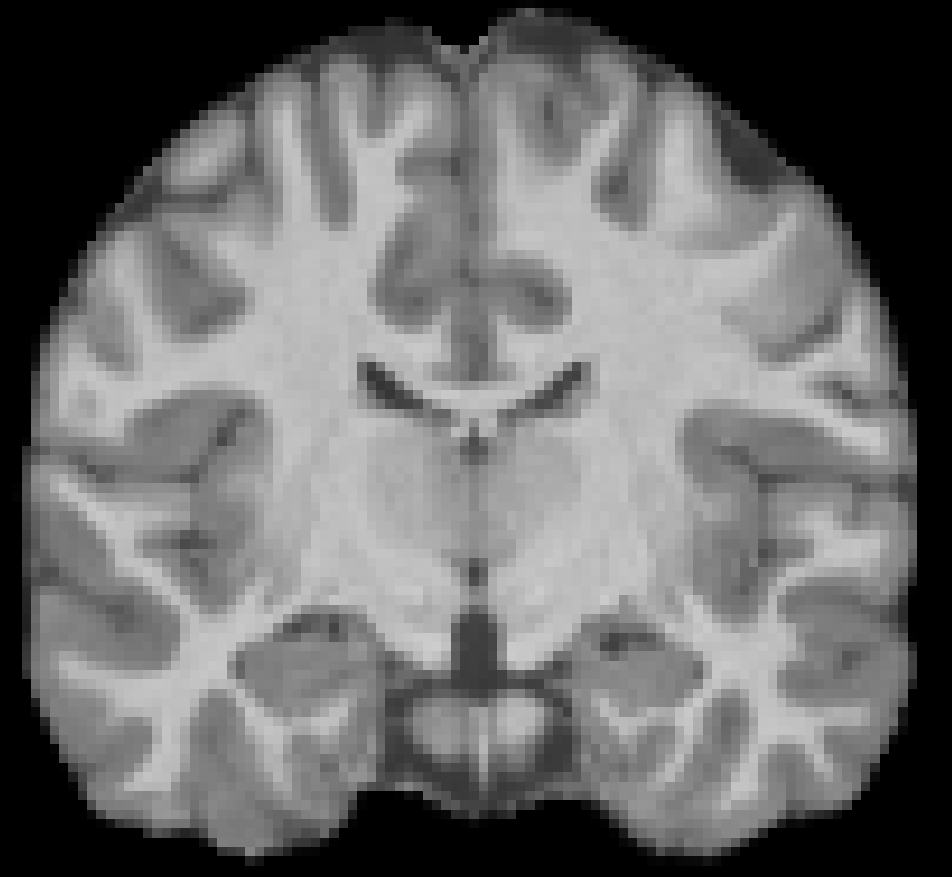
Age 18 F, NKI-RS Cognitively normal |
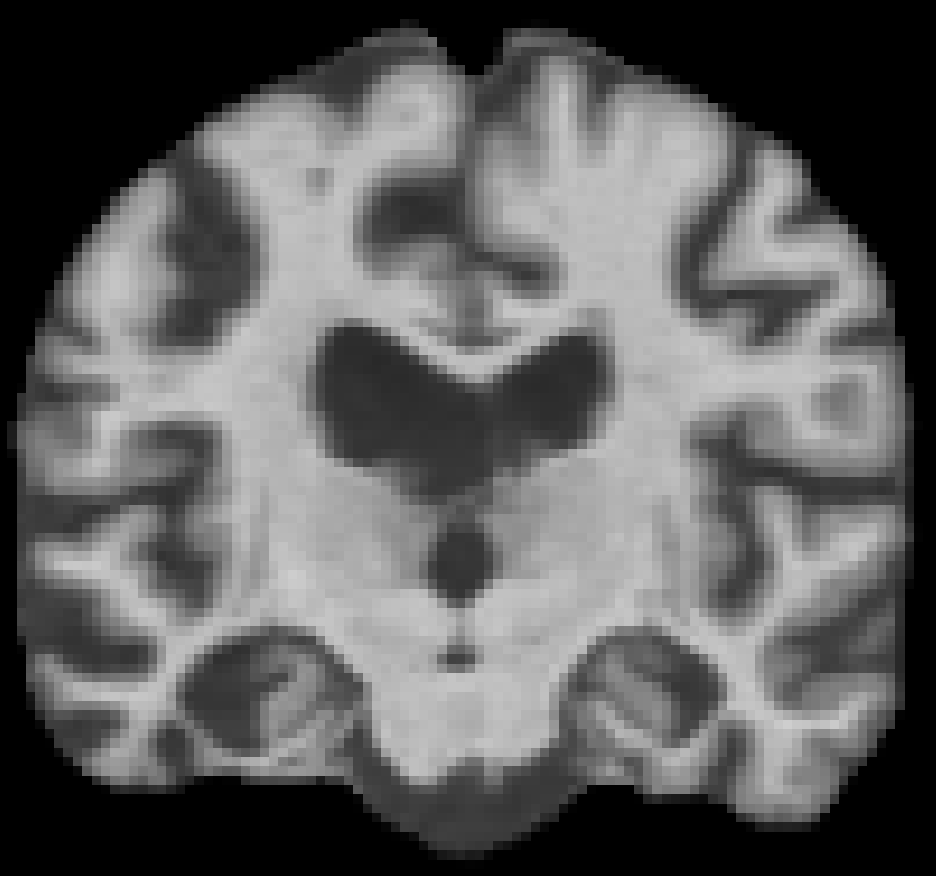
Age 71 M, OASIS-1 Alzheimer's disease |
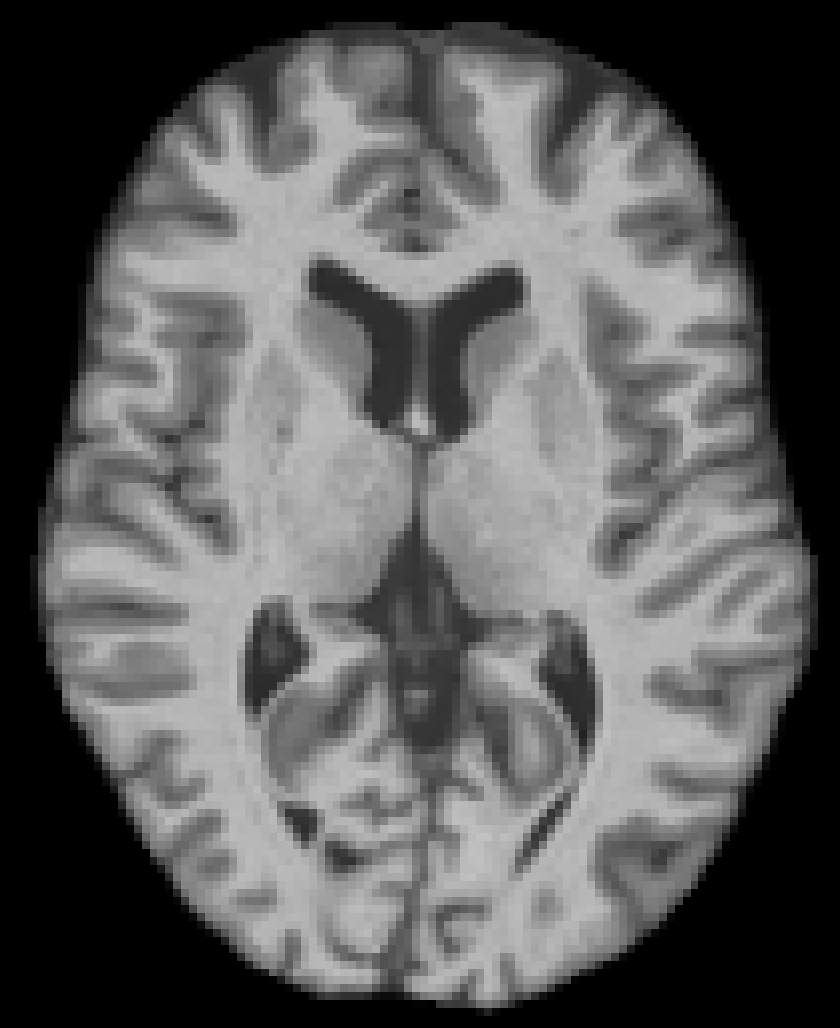
Age 46 F, IXI Cognitively normal |
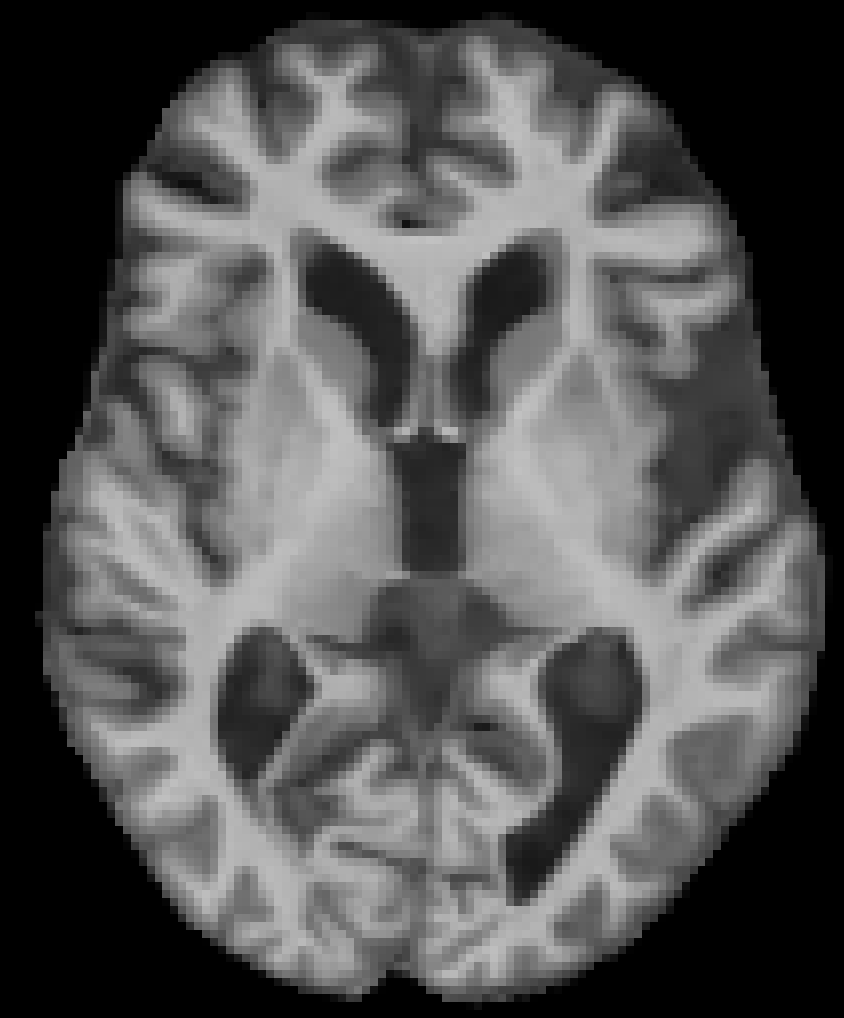
Age 86 M, OASIS-2 Cognitively normal |
Subject characteristics table
| Split | n (scans) | n (subjects) | age_mean | age_std | age_range | sex_counts | diagnosis_counts | study_counts |
|---|---|---|---|---|---|---|---|---|
| train | 3066 | 2085 | 45.1 | 24.5 | (6, 98) | {'female': 1827, 'male': 1239} | {'CN': 2847, 'AD': 219} | {'NKI-RS': 1854, 'OASIS-1': 340, 'IXI': 326, 'OASIS-2': 296, 'DLBS': 250} |
| validation | 364 | 261 | 46.4 | 24.5 | (6, 90) | {'female': 225, 'male': 139} | {'CN': 339, 'AD': 25} | {'NKI-RS': 213, 'IXI': 43, 'OASIS-1': 38, 'OASIS-2': 38, 'DLBS': 32} |
| test | 364 | 261 | 45.7 | 24.6 | (6, 93) | {'female': 210, 'male': 154} | {'CN': 343, 'AD': 21} | {'NKI-RS': 216, 'IXI': 40, 'OASIS-2': 39, 'OASIS-1': 36, 'DLBS': 33} |
Folder organization
brain-structure/
├─ brain-structure.py
├─ metadata.csv
├─ IXI/
│ ├─ sub-002/
│ │ └─ ses-01/
│ │ └─ anat/
│ │ ├─ msub-002_ses-01_T1w_brain_affine_mni.nii.gz
│ │ └─ msub-002_ses-01_scandata.json
│ └─ ...
├─ DLBS/
│ └─ ...
└─ ...
Example usage
# install Hugging Face Datasets
pip install datasets
# optional installs: NiBabel and PyTorch
pip install nibabel
pip install torch torchvision
# load datasets
from datasets import load_dataset
ds_train = load_dataset("radiata-ai/brain-structure", split="train", trust_remote_code=True)
ds_val = load_dataset("radiata-ai/brain-structure", split="validation", trust_remote_code=True)
ds_test = load_dataset("radiata-ai/brain-structure", split="test", trust_remote_code=True)
# example PyTorch processing of images
import nibabel as nib
import torch
import torch.nn.functional as F
from torch.utils.data import Dataset
def preprocess_nifti(example):
"""
Loads a .nii.gz file, crops, normalizes, and resamples to 96^3.
Returns a numpy array (or tensor) in example["img"].
"""
nii_path = example["nii_filepath"]
# Load volume data
vol = nib.load(nii_path).get_fdata()
# Crop sub-volume
vol = vol[7:105, 8:132, :108] # shape: (98, 124, 108)
# Shift intensities to be non-negative
vol = vol + abs(vol.min())
# Normalize to [0,1]
vol = vol / vol.max()
# Convert to torch.Tensor: (1,1,D,H,W)
t_tensor = torch.from_numpy(vol).float().unsqueeze(0).unsqueeze(0)
# Scale factor based on (124 -> 96) for the y-dimension
scale_factor = 96 / 124
downsampled = F.interpolate(
t_tensor,
scale_factor=(scale_factor, scale_factor, scale_factor),
mode="trilinear",
align_corners=False
)
# Now pad each dimension to exactly 96 (symmetric padding)
_, _, d, h, w = downsampled.shape
pad_d = 96 - d
pad_h = 96 - h
pad_w = 96 - w
padding = (
pad_w // 2, pad_w - pad_w // 2,
pad_h // 2, pad_h - pad_h // 2,
pad_d // 2, pad_d - pad_d // 2
)
final_img = F.pad(downsampled, padding) # shape => (1, 1, 96, 96, 96)
final_img = final_img.squeeze(0)
# Store as numpy or keep as torch.Tensor
example["img"] = final_img.numpy()
return example
# Apply the preprocessing to each split
ds_train = ds_train.map(preprocess_nifti)
ds_val = ds_val.map(preprocess_nifti)
ds_test = ds_test.map(preprocess_nifti)
# Set the dataset format to return PyTorch tensors for the 'img' column
ds_train.set_format(type='torch', columns=['img'])
ds_val.set_format(type='torch', columns=['img'])
ds_test.set_format(type='torch', columns=['img'])
# Set up data loaders for model training
train_loader = DataLoader(ds_train, batch_size=16, shuffle=True)
val_loader = DataLoader(ds_val, batch_size=16, shuffle=False)
test_loader = DataLoader(ds_test, batch_size=16, shuffle=False)
Study descriptions
- IXI: A dataset of nearly 600 MR images from normal, healthy subjects, including T1, T2, PD-weighted, MRA, and diffusion-weighted images collected at three different hospitals in London.
Citation: IXI data was obtained from https://brain-development.org/ixi-dataset/ - DLBS: A dataset from the Dallas Lifespan Brain Study (DLBS) comprising structural MRI, DTI, functional MRI, resting-state fMRI, and amyloid PET scans from 350 healthy adults aged 20-89, including extensive cognitive testing and demographic information.
Citation: DLBS data was obtained from the International Neuroimaging Data-sharing Initiative (INDI) database. - NKI-RS: A large-scale ongoing neuroimaging dataset (N > 1000) across the lifespan from a community sample, including structural and functional MRI scans such as MPRAGE, DTI, resting-state fMRI, and task-based fMRI.
Citation: NKI-RS data was obtained from Rockland Sample Neuroimaging Data Release. - OASIS-1: Cross-sectional T1-weighted MRI data from 416 right-handed subjects aged 18 to 96, including 100 over 60 with very mild to moderate Alzheimer’s disease, each with 3 or 4 scans.
Citation: OASIS-1: Cross-Sectional: https://doi.org/10.1162/jocn.2007.19.9.1498 - OASIS-2: A longitudinal MRI dataset of 150 right-handed individuals aged 60-96, with 373 imaging sessions including T1-weighted MRIs, featuring nondemented and demented older adults, including patients with Alzheimer’s disease.
Citation: OASIS-2: Longitudinal: https://doi.org/10.1162/jocn.2009.21407
Methods
Image processing
T1-weighted structural MRI scans were processed with CAT12 (Gaser et al, 2024). The image processing steps were:
- correct for bias, noise, and intensity
- mask to brain-only (gray matter + white matter + CSF)
- register to ICBM 2009c Nonlinear Asymmetric space (MNI152NLin2009cAsym 1.5mm^3) using linear affine registration with 12 degrees of freedom in FSL FLIRT ('flirt -in t1.nii.gz -ref mni_icbm152_t1_tal_nlin_asym_09c_brain_1_5_mm.nii.gz -dof 12 -noresampblur').
The goal was to get denoised, unsmoothed scans that were maximally aligned to standard space while preserving individual anatomy.
Metadata includes the total intracranial volume (TIV), image quality rating (IQR; larger value = worse quality), MRI scanner manufacturer/model, and field strength.
Train/validation/test partitioning
Scans were partitioned into train/validation/test datasets with a 80%/10%/10% split. Splits were balanced for age, sex, clinical diagnosis, and study. Subjects with multiple scans only appear in one split.
Citation
@dataset{Radiata-Brain-Structure,
author = {Jesse Brown and Clayton Young},
title = {Brain-Structure: Processed Structural MRI Brain Scans Across the Lifespan},
year = {2025},
url = {https://huggingface.co/datasets/radiata-ai/brain-structure},
note = {Version 1.0},
publisher = {Hugging Face}
}
- Downloads last month
- 487