license: apache-2.0
datasets:
- FreedomIntelligence/ApolloMoEDataset
language:
- ar
- en
- zh
- ko
- ja
- mn
- th
- vi
- lo
- mg
- de
- pt
- es
- fr
- ru
- it
- hr
- gl
- cs
- co
- la
- uk
- bs
- bg
- eo
- sq
- da
- sa
- 'no'
- gn
- sr
- sk
- gd
- lb
- hi
- ku
- mt
- he
- ln
- bm
- sw
- ig
- rw
- ha
metrics:
- accuracy
base_model:
- microsoft/Phi-3-mini-4k-instruct
pipeline_tag: question-answering
tags:
- biology
- medical
Covering 12 Major Languages including English, Chinese, French, Hindi, Spanish, Arabic, Russian, Japanese, Korean, German, Italian, Portuguese and 38 Minor Languages So far.
📃 Paper • 🌐 Demo • 🤗 ApolloMoEDataset • 🤗 ApolloMoEBench • 🤗 Models • 🌐 Apollo • 🌐 ApolloMoE
🌈 Update
- [2024.10.15] ApolloMoE repo is published!🎉
Architecture
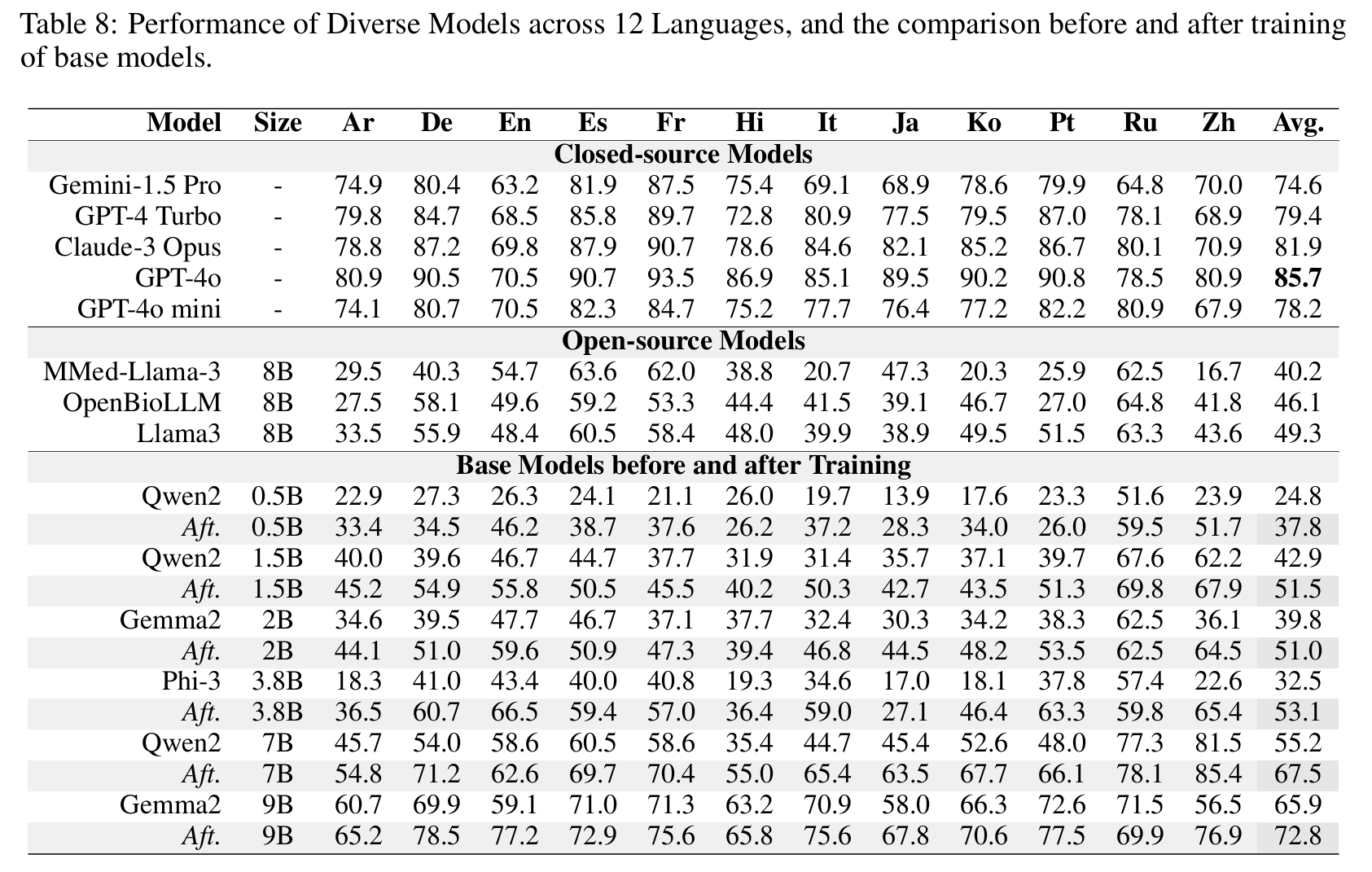
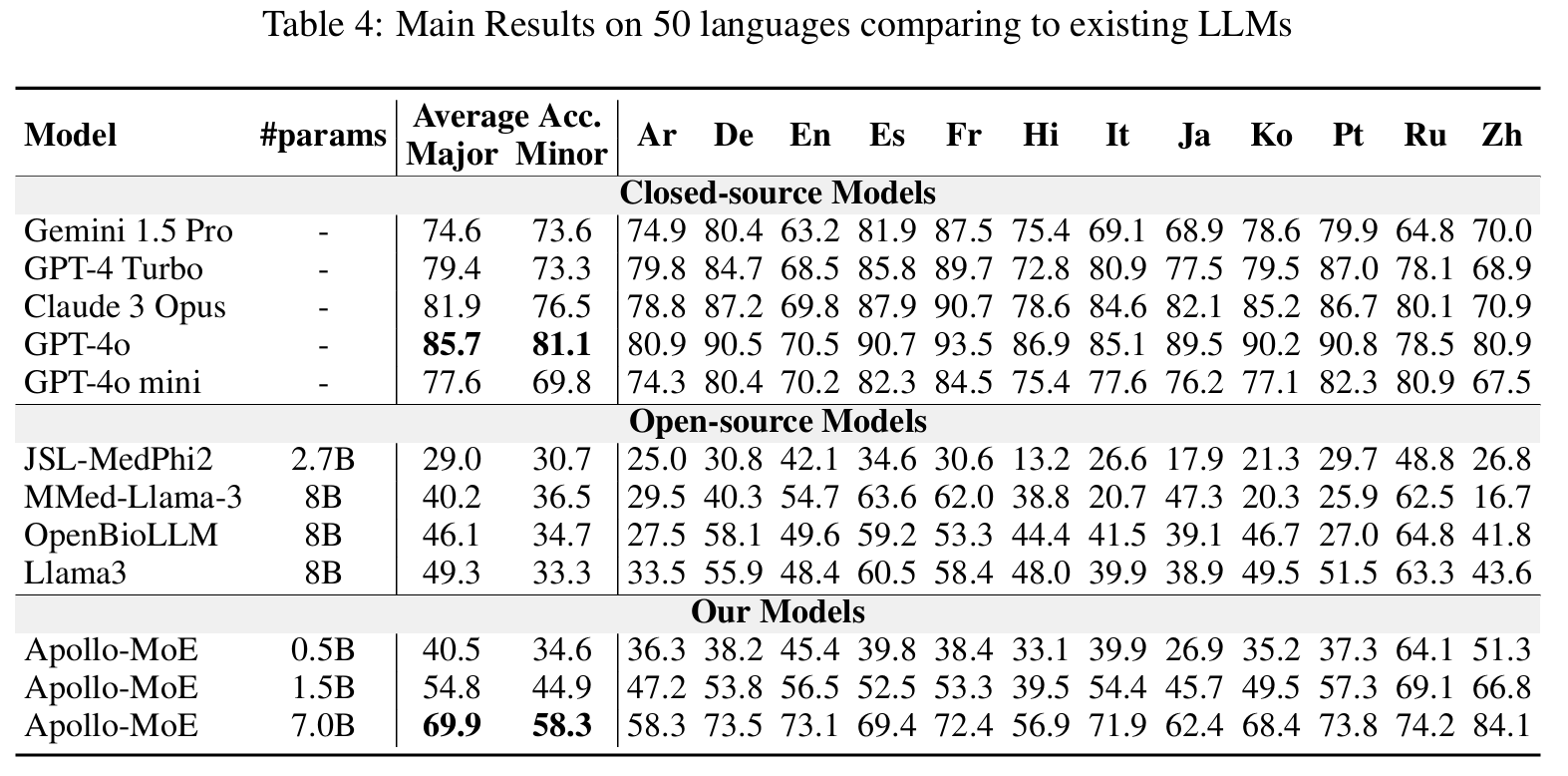
Results
Dense
🤗 Apollo2-0.5B • 🤗 Apollo2-1.5B • 🤗 Apollo2-2B • 🤗 Apollo2-3.8B • 🤗 Apollo2-7B • 🤗 Apollo2-9B
Post-MoE
🤗 Apollo-MoE-0.5B • 🤗 Apollo-MoE-1.5B • 🤗 Apollo-MoE-7B
Usage Format
Apollo2
- 0.5B, 1.5B, 7B: User:{query}\nAssistant:{response}<|endoftext|>
- 2B, 9B: User:{query}\nAssistant:{response}<eos>
- 3.8B: <|user|>\n{query}<|end|><|assisitant|>\n{response}<|end|>
Apollo-MoE
- 0.5B, 1.5B, 7B: User:{query}\nAssistant:{response}<|endoftext|>
Dataset & Evaluation
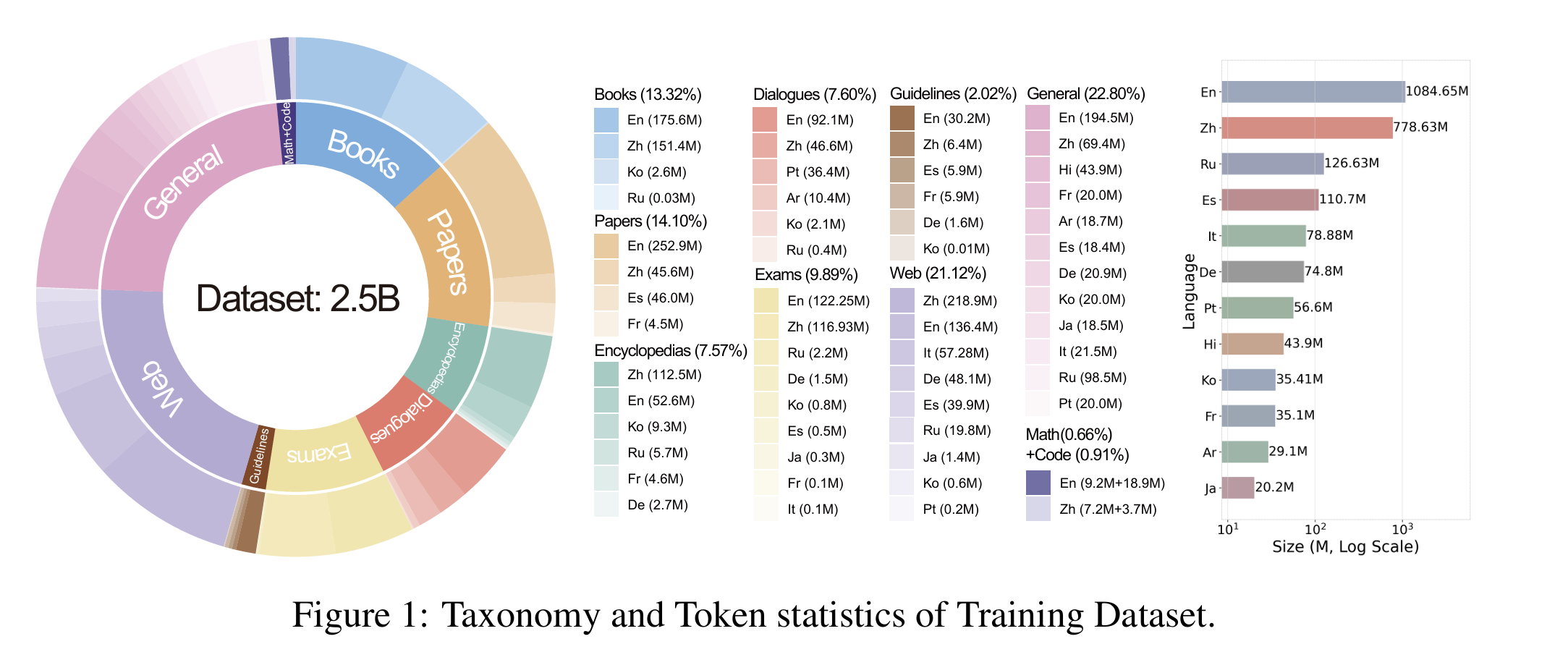
Dataset 🤗 ApolloMoEDataset
Evaluation 🤗 ApolloMoEBench
Click to expand
EN:
- MedQA-USMLE
- MedMCQA
- PubMedQA: Because the results fluctuated too much, they were not used in the paper.
- MMLU-Medical
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
ZH:
- MedQA-MCMLE
- CMB-single: Not used in the paper
- Randomly sample 2,000 multiple-choice questions with single answer.
- CMMLU-Medical
- Anatomy, Clinical_knowledge, College_medicine, Genetics, Nutrition, Traditional_chinese_medicine, Virology
- CExam: Not used in the paper
- Randomly sample 2,000 multiple-choice questions
ES: Head_qa
FR:
- Frenchmedmcqa
- [MMLU_FR]
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
HI: MMLU_HI
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
AR: MMLU_AR
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
JA: IgakuQA
KO: KorMedMCQA
IT:
- MedExpQA
- [MMLU_IT]
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
DE: BioInstructQA: German part
PT: BioInstructQA: Portuguese part
RU: RuMedBench
Results reproduction
Click to expand
We take Gemma-2b as example
Download Dataset for project:
bash 0.download_data.shPrepare test and dev for specific model:
- Create test data for with special token, you can use ./util/check.ipynb to check models' special tokens
bash 1.data_process_test&dev.shPrepare train data for specific model (Create tokenized data in advance):
- You can adjust data Training order and Training Epoch in this step
bash 2.data_process_train.shTrain the model
- If you want to train in Multi Nodes please refer to ./scripts/multi_node_train_*.sh
bash 3.single_node_train_gemma.shEvaluate your model: Generate score for benchmark
bash 4.eval.shEvaluate your model: Play with your ckpts in bash
python ./src/evaluate/cli_demo.py --model_name='./ckpts/your/path/tfmr'
Citation
Please use the following citation if you intend to use our dataset for training or evaluation:
@misc{zheng2024efficientlydemocratizingmedicalllms,
title={Efficiently Democratizing Medical LLMs for 50 Languages via a Mixture of Language Family Experts},
author={Guorui Zheng and Xidong Wang and Juhao Liang and Nuo Chen and Yuping Zheng and Benyou Wang},
year={2024},
eprint={2410.10626},
archivePrefix={arXiv},
primaryClass={cs.CL},
url={https://arxiv.org/abs/2410.10626},
}