|
--- |
|
license: mit |
|
--- |
|
--- |
|
license: mit |
|
tags: |
|
- vision |
|
- image-segmentation |
|
- instance-segmentation |
|
datasets: |
|
- custom-germination-dataset |
|
widget: |
|
- src: https://example.com/path/to/germination-image1.jpg |
|
example_title: Germination Image 1 |
|
- src: https://example.com/path/to/germination-image2.jpg |
|
example_title: Germination Image 2 |
|
--- |
|
|
|
# GermiNet: A MaskFormer Model for Germination Counting |
|
|
|
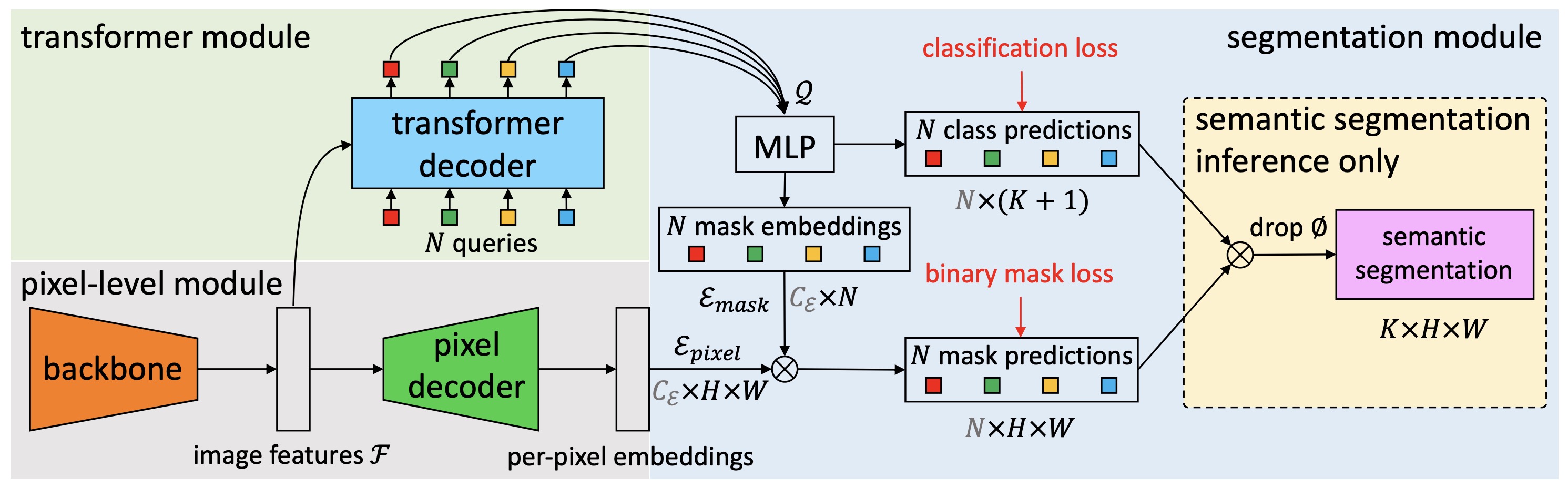
GermiNet model trained on a custom germination dataset for instance segmentation (small-sized version, Swin backbone). It is based on the MaskFormer architecture introduced in the paper [MaskFormer: Per-Pixel Classification is Not All You Need for Semantic Segmentation](https://arxiv.org/abs/2107.06278) and first released in [this repository](https://github.com/facebookresearch/MaskFormer). |
|
|
|
Disclaimer: This model card is written by [Your Name/Organization] with assistance from Grok (xAI) for the Hugging Face community. |
|
|
|
## Model Description |
|
|
|
GermiNet is a MaskFormer-based instance segmentation model fine-tuned to detect and segment "normal" and "abnormal" seeds in germination images. It uses the `facebook/maskformer-swin-tiny-coco` pre-trained checkpoint as its backbone, with a Swin-Tiny transformer architecture. The model predicts a set of masks and corresponding labels for three classes: "background," "normal," and "abnormal," with an additional "no object" class handled internally by MaskFormer. The model was trained on a small custom dataset as a proof-of-concept for automating germination counting in agricultural research. |
|
|
|
 |
|
|
|
## Intended Uses & Limitations |
|
|
|
GermiNet is intended for instance segmentation tasks in agricultural research, specifically for detecting and segmenting "normal" and "abnormal" seeds in germination images. It can be used with tools like CVAT for automated annotation workflows. |
|
|
|
### Limitations |
|
- The model was trained on a small dataset (18 images), which limits its generalization. |
|
- Local inference shows a bias toward "no object" predictions, with few "normal" detections and no "abnormal" detections, indicating underfitting. |
|
- Mask resolution is 56x56 (upscaled to 224x224 or higher for visualization), which may miss fine details. |
|
- The model requires further training with a larger dataset and more epochs for improved performance. |
|
|
|
See the [model hub](https://huggingface.co/models?search=germinet) to look for other fine-tuned versions if needed. |
|
|
|
## How to Use |
|
|
|
Here’s how to use this model for instance segmentation: |
|
|
|
```python |
|
import requests |
|
import torch |
|
from PIL import Image |
|
from transformers import AutoImageProcessor, MaskFormerForInstanceSegmentation |
|
|
|
# Load GermiNet fine-tuned on custom germination dataset |
|
processor = AutoImageProcessor.from_pretrained("your-username/germi-net") |
|
model = MaskFormerForInstanceSegmentation.from_pretrained("your-username/germi-net") |
|
|
|
# Load an image (replace with your image URL or local path) |
|
url = "https://example.com/path/to/germination-image.jpg" |
|
image = Image.open(requests.get(url, stream=True).raw) |
|
# Alternatively, use a local image |
|
# image = Image.open("path/to/your/image.jpg") |
|
inputs = processor(images=image, return_tensors="pt") |
|
|
|
# Run inference |
|
with torch.no_grad(): |
|
outputs = model(**inputs) |
|
|
|
# Model predicts class_queries_logits and masks_queries_logits |
|
class_queries_logits = outputs.class_queries_logits # Shape: (batch_size, num_queries, num_classes + 1) |
|
masks_queries_logits = outputs.masks_queries_logits # Shape: (batch_size, num_queries, height, width) |
|
|
|
# Post-process predictions |
|
predicted_classes = class_queries_logits.argmax(-1).cpu().numpy() |
|
mask_predictions = masks_queries_logits.sigmoid().cpu().numpy() |
|
binary_masks = (mask_predictions > 0.5).astype(np.uint8) |
|
|
|
# Map predictions to labels |
|
id2label = {0: "background", 1: "normal", 2: "abnormal", 3: "no object"} |
|
predicted_labels = [id2label[cls] for cls in predicted_classes[0]] |
|
print("Predicted labels:", predicted_labels) |
|
|
|
# Optional: Visualize (requires matplotlib and cv2) |
|
import numpy as np |
|
import matplotlib.pyplot as plt |
|
import cv2 |
|
|
|
visualization_size = (800, 800) |
|
resized_masks = np.zeros((binary_masks.shape[1], *visualization_size), dtype=np.uint8) |
|
for i in range(binary_masks.shape[1]): |
|
resized_masks[i] = cv2.resize(binary_masks[0, i], visualization_size, interpolation=cv2.INTER_NEAREST) |
|
|
|
image_np = np.array(image) |
|
aspect_ratio = image_np.shape[1] / image_np.shape[0] |
|
new_height = visualization_size[0] |
|
new_width = int(new_height * aspect_ratio) |
|
resized_image = cv2.resize(image_np, (new_width, new_height), interpolation=cv2.INTER_LINEAR) |
|
if new_width != visualization_size[1]: |
|
start_x = (new_width - visualization_size[1]) // 2 |
|
resized_image = resized_image[:, start_x:start_x + visualization_size[1]] |