fixed READMEs and added IDR Prediction benchmark
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- fuson_plm/benchmarking/caid/README.md +44 -42
- fuson_plm/benchmarking/idr_prediction/README.md +211 -0
- fuson_plm/benchmarking/idr_prediction/__init__.py +0 -0
- fuson_plm/benchmarking/idr_prediction/clean.py +289 -0
- fuson_plm/benchmarking/idr_prediction/cluster.py +94 -0
- fuson_plm/benchmarking/idr_prediction/clustering/input.fasta +3 -0
- fuson_plm/benchmarking/idr_prediction/clustering/mmseqs_full_results.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/config.py +41 -0
- fuson_plm/benchmarking/idr_prediction/model.py +154 -0
- fuson_plm/benchmarking/idr_prediction/plot.py +204 -0
- fuson_plm/benchmarking/idr_prediction/processed_data/all_albatross_seqs_and_properties.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/processed_data/train_test_value_histograms.png +0 -0
- fuson_plm/benchmarking/idr_prediction/processed_data/value_histograms.png +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/asph_bio_synth_training_data_cleaned_05_09_2023.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/asph_nat_meth_test.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/scaled_re_bio_synth_training_data_cleaned_05_09_2023.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/scaled_re_nat_meth_test.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/scaled_rg_bio_synth_training_data_cleaned_05_09_2023.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/scaled_rg_nat_meth_test.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/scaling_exp_bio_synth_training_data_cleaned_05_09_2023.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/raw_data/scaling_exp_nat_meth_test.tsv +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/asph_best_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/asph_hyperparam_screen_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/esm2_t33_650M_UR50D_asph_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/esm2_t33_650M_UR50D_asph_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/fuson_plm_asph_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/fuson_plm_asph_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/esm2_t33_650M_UR50D_scaled_re_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/esm2_t33_650M_UR50D_scaled_re_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/fuson_plm_scaled_re_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/fuson_plm_scaled_re_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/esm2_t33_650M_UR50D_scaled_rg_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/esm2_t33_650M_UR50D_scaled_rg_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/fuson_plm_scaled_rg_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/fuson_plm_scaled_rg_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/esm2_t33_650M_UR50D_scaling_exp_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/esm2_t33_650M_UR50D_scaling_exp_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/fuson_plm_scaling_exp_R2.png +0 -0
- fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/fuson_plm_scaling_exp_R2_source_data.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/scaled_re_best_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/scaled_re_hyperparam_screen_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/scaled_rg_best_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/scaled_rg_hyperparam_screen_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/scaling_exp_best_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/results/final/scaling_exp_hyperparam_screen_test_r2.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/split.py +135 -0
- fuson_plm/benchmarking/idr_prediction/splits/asph/test_df.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/splits/asph/train_df.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/splits/asph/val_df.csv +3 -0
- fuson_plm/benchmarking/idr_prediction/splits/scaled_re/test_df.csv +3 -0
fuson_plm/benchmarking/caid/README.md
CHANGED
|
@@ -120,47 +120,46 @@ benchmarking/
|
|
| 120 |
|
| 121 |
Here we describe what each script does and which files each script creates.
|
| 122 |
1. 🐍 **`scrape_fusionpdb.py`**
|
| 123 |
-
|
| 124 |
-
|
| 125 |
-
|
| 126 |
-
|
| 127 |
-
|
| 128 |
-
|
| 129 |
-
|
| 130 |
-
|
| 131 |
-
|
| 132 |
-
|
| 133 |
-
|
| 134 |
-
|
| 135 |
-
|
| 136 |
-
|
| 137 |
-
|
| 138 |
-
|
| 139 |
-
|
| 140 |
-
|
| 141 |
-
|
| 142 |
-
|
| 143 |
-
|
| 144 |
-
|
| 145 |
-
|
| 146 |
-
|
| 147 |
-
<br>
|
| 148 |
|
| 149 |
2. 🐍 **`process_fusion_structures.py`**
|
| 150 |
-
|
| 151 |
-
|
| 152 |
-
|
| 153 |
-
|
| 154 |
-
|
| 155 |
-
|
| 156 |
-
|
| 157 |
-
|
| 158 |
-
|
| 159 |
-
|
| 160 |
-
|
| 161 |
-
|
| 162 |
-
|
| 163 |
-
|
| 164 |
|
| 165 |
|
| 166 |
### Training
|
|
@@ -184,9 +183,12 @@ PERMISSION_TO_OVERWRITE_EMBEDDINGS = False # if False, scrip
|
|
| 184 |
PERMISSION_TO_OVERWRITE_MODELS = False # if False, script will halt if it believes these embeddings have already been made.
|
| 185 |
```
|
| 186 |
|
| 187 |
-
|
|
|
|
|
|
|
|
|
|
| 188 |
|
| 189 |
-
|
| 190 |
|
| 191 |
```
|
| 192 |
benchmarking/
|
|
@@ -209,7 +211,7 @@ benchmarking/
|
|
| 209 |
- **`caid_train_losses.csv`**: train losses over the 2 training epochs for top-performing model
|
| 210 |
- **`params.txt`**: hyperparameters of top performing model
|
| 211 |
|
| 212 |
-
|
| 213 |
|
| 214 |
```
|
| 215 |
benchmarking/
|
|
|
|
| 120 |
|
| 121 |
Here we describe what each script does and which files each script creates.
|
| 122 |
1. 🐍 **`scrape_fusionpdb.py`**
|
| 123 |
+
i. Scrapes metadata for FusionPDB Level 2 and Level 3
|
| 124 |
+
a. Pulls the online tables for [Level 2](https://compbio.uth.edu/FusionPDB/gene_search_result_0.cgi?type=chooseLevel&chooseLevel=level2) and [Level 3](https://compbio.uth.edu/FusionPDB/gene_search_result_0.cgi?type=chooseLevel&chooseLevel=level3), saving results to `raw_data/FusionPDB_level2_curated_09_05_2024.csv` and `raw_data/FusionPDB_level3_curated_09_05_2024.csv` respectively.
|
| 125 |
+
ii. Retrieves structure links
|
| 126 |
+
a. Using the tables collected in step (i), visits the page for each fusion oncoprotein (FO) in FusionPDB Level 2 and 3, and downloads all AlphaFold2 structure links for each FO.
|
| 127 |
+
b. Saves results directly to `raw_data/FusionPDB_level2_fusion_structure_links.csv` and `raw_data/FusionPDB_level3_fusion_structure_links.csv`, respectively
|
| 128 |
+
iii. Retrieves FO head gene and tail gene info
|
| 129 |
+
a. Using the tables collected in step (i), visits the page for each fusion oncoprotein (FO) in FusionPDB Level 2 and 3 to download head/tail info. Collects HGID and TGID (GeneIDs for head and tail) and UniProt accessions for each.
|
| 130 |
+
b. Saves results directly to `raw_data/level2_head_tail_info.txt` and `raw_data/level3_head_tail_info.txt`, respectively.
|
| 131 |
+
iv. Combines Level 2 and 3 head/tail data
|
| 132 |
+
a. Merges `raw_data/level2_head_tail_info.txt` and `raw_data/level3_head_tail_info.txt` into a dataframe.
|
| 133 |
+
b. Saves result at `processed_data/fusionpdb/fusion_heads_and_tails.csv` (columns="FusionGID","HGID","TGID","HGUniProtAcc","TGUniProtAcc")
|
| 134 |
+
v. Combines Level 2 and 3 structure link data
|
| 135 |
+
a. Joins structure link data with metadata for each of levels 2 and 3, then combines the result.
|
| 136 |
+
b. Saves result at `processed_data/fusionpdb/intermediates/giant_level2-3_fusion_protein_structure_links.csv`
|
| 137 |
+
vi. Combines structure link data and metadata (result of step (v)) with head and tail data (result of step (iv)), and resolves any missing head/tail UniProt IDs.
|
| 138 |
+
a. Merges the data
|
| 139 |
+
b. Checks how many rows have either missing or wrong UniProt accessions for the head or tail gene, and compiles the gene symbols for online quering in the UniProt ID Mapping tool (`processed_data/fusionpdb/intermediates/unmapped_parts.txt`)
|
| 140 |
+
c. Reads the UniProt ID Mapping result. Combines this data with FusionPDB-scraped data by matching FusionPDB's HGID (GeneID for head) and TGID (GeneID for tail) with the GeneID returned by UniProt.
|
| 141 |
+
d. For any FO where FusionPDB lacked a UniProt ID for the head/tail, this ID is filled in from the UniProt ID Mapping result.
|
| 142 |
+
e. Saves result to `processed_data/fusionpdb/intermediates/giant_level2-3_fusion_protein_head_tail_info.csv`. Columns: "FusionGID","FusionGene","Hgene","Tgene","URL","HGID","TGID","HGUniProtAcc","TGUniProtAcc","HGUniProtAcc_Source","TGUniProtAcc_Source", where the "_Source" columns indicate whether the UniProt ID came from FusionPDB, or from the ID Map.
|
| 143 |
+
vii. Downloads AlphaFold2 structures of FOs from FusionPDB.
|
| 144 |
+
a. Using structure links from `processed_data/fusionpdb/intermediates/giant_level2-3_fusion_protein_structure_links.csv` (step (v)), directly downloads `.pdb` and `.cif` files.
|
| 145 |
+
b. Saves results in 📁`raw_data/fusionpdb/structures`
|
| 146 |
+
|
|
|
|
| 147 |
|
| 148 |
2. 🐍 **`process_fusion_structures.py`**
|
| 149 |
+
i. Determines pLDDT(s) for each FO structure.
|
| 150 |
+
a. For each structure in 📁`raw_data/fusionpdb_structures/`, determines amino acid sequence, per-residue pLDDT, and average pLDDT from the AlphaFold2 structure.
|
| 151 |
+
b. Saves results in `processed_data/fusionpdb/intermediates/giant_level2-3_fusion_protein_structures_processed.csv`.
|
| 152 |
+
ii. Downloads AlphaFold2 structures for all head and tail proteins
|
| 153 |
+
a. Reads `processed_data/fusionpdb/intermediates/giant_level2-3_fusion_protein_head_tail_info.csv` and collects all unique UniProt IDs for all head/tail proteins.
|
| 154 |
+
b. For each UniProt ID, queries the AlphaFoldDB, downloads the AlphaFold2 structure (if available), and saves it to 📁`raw_data/fusionpdb/head_tail_af2db_structures/`. Saves files converted from PDB to CIF format in `mmcif_converted_files`. Then, extracts the sequence, per-residue pLDDT, and average pLDDT from the file.
|
| 155 |
+
c. Saves any UniProt IDs that did not have structures in the AlphaFoldDB to: `processed_data/fusionpdb/intermediates/uniprotids_not_in_afdb.txt`. Most of these were very long, but the shorter ones were folded and their average pLDDTs were manually inputted. These were put back into the AlphaFold ID map to look for alternative UniProt IDs, and their results are in `not_in_afdb_idmap.txt`.
|
| 156 |
+
d. Saves results to `processed_data/fusionpdb/heads_tails_structural_data.csv`
|
| 157 |
+
iii. Cleans the dataase of level 2&3 structural info
|
| 158 |
+
a. Drops rows where no structure was successfully downloaded
|
| 159 |
+
b. Drops rows where the FO sequence from FusionPDB does not match the FO sequence from its own AlphaFold2 structure file
|
| 160 |
+
c. ⭐️Saves **two final, cleaned databases**⭐️:
|
| 161 |
+
a. ⭐️ **`FusionPDB_level2-3_cleaned_FusionGID_info.csv`**: includes ful IDs and structural information for the Hgene and Tgene of each FO. Columns="FusionGID","FusionGene","Hgene","Tgene","URL","HGID","TGID","HGUniProtAcc","TGUniProtAcc","HGUniProtAcc_Source","TGUniProtAcc_Source","HG_pLDDT","HG_AA_pLDDTs","HG_Seq","TG_pLDDT","TG_AA_pLDDTs","TG_Seq".
|
| 162 |
+
b. ⭐️ **`FusionPDB_level2-3_cleaned_structure_info.csv`**: includes full structural information for each FO. Columns= "FusionGID","FusionGene","Fusion_Seq","Fusion_Length","Hgene","Hchr","Hbp","Hstrand","Tgene","Tchr","Tbp","Tstrand","Level","Fusion_Structure_Link","Fusion_Structure_Type","Fusion_pLDDT","Fusion_AA_pLDDTs","Fusion_Seq_Source"
|
| 163 |
|
| 164 |
|
| 165 |
### Training
|
|
|
|
| 183 |
PERMISSION_TO_OVERWRITE_MODELS = False # if False, script will halt if it believes these embeddings have already been made.
|
| 184 |
```
|
| 185 |
|
| 186 |
+
`train.py` trains the models using embeddings indicated in `config.py`. It also performs a hyperparameter screen.
|
| 187 |
+
- All **results** are stored in `caid/results/<timestamp>`, where `timestamp` is a unique string encoding the date and time when you started training.
|
| 188 |
+
- All **raw outputs from models** are stored in `caid/trained_models/<embedding_path>`, where `embedding_path` represents the embeddings used to build the disorder predictor.
|
| 189 |
+
- All **embeddings** made for training will be stored in a new folder called `caid/embeddings/` with subfolders for each model. This allows you to use the same model multiple times without regenerating embeddings.
|
| 190 |
|
| 191 |
+
Below is the FusOn-pLM-Diso raw outputs folder, `trained_models/fuson_plm/best/'. (ESM-2-650M-Diso has a folder in the same format, and future trained models will as well):
|
| 192 |
|
| 193 |
```
|
| 194 |
benchmarking/
|
|
|
|
| 211 |
- **`caid_train_losses.csv`**: train losses over the 2 training epochs for top-performing model
|
| 212 |
- **`params.txt`**: hyperparameters of top performing model
|
| 213 |
|
| 214 |
+
Results from the FusOn-pLM manuscript are found in `results/final`. A few extra data files and plots are added by `analyze_fusion_preds.py`
|
| 215 |
|
| 216 |
```
|
| 217 |
benchmarking/
|
fuson_plm/benchmarking/idr_prediction/README.md
ADDED
|
@@ -0,0 +1,211 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
## IDR Property Prediction Benchmark
|
| 2 |
+
|
| 3 |
+
This folder contains all the data and code needed to perform the **IDR property prediction benchmark**, where FusOn-pLM-IDR (a regressor built on FusOn-pLM embeddings) is used to predict aggregate properties of intrinsically disordered regions (IDRs), specifically asphericity, end-to-end radius (R<sub>e</sub>), radius of gyration (R<sub>g</sub>), and polymer scaling exponent (Figure 4A-B).
|
| 4 |
+
|
| 5 |
+
### TL;DR
|
| 6 |
+
The order in which to run the scripts, after downloading data:
|
| 7 |
+
|
| 8 |
+
```
|
| 9 |
+
python clean.py # clean the data
|
| 10 |
+
python cluster.py # MMSeqs2 clustering
|
| 11 |
+
python split.py # make cluster-based train/val/test splits
|
| 12 |
+
python train.py # train the model
|
| 13 |
+
python plot.py # if you want to remake r2 plots
|
| 14 |
+
```
|
| 15 |
+
|
| 16 |
+
### Downloading raw IDR data
|
| 17 |
+
IDR properties from [Lotthammer et al. 2024](https://doi.org/10.1038/s41592-023-02159-5) (ALBATROSS model) were used to train FusOn-pLM-Diso. Sequences were downloaded from [this link](https://github.com/holehouse-lab/supportingdata/blob/master/2023/ALBATROSS_2023/simulations/data/all_sequences.tgz) and deposited in `raw_data`. All files in `raw_data` are from this direct download.
|
| 18 |
+
|
| 19 |
+
```
|
| 20 |
+
benchmarking/
|
| 21 |
+
└── idr_prediction/
|
| 22 |
+
└── raw_data/
|
| 23 |
+
├── asph_bio_synth_training_data_cleaned_05_09_2023.tsv
|
| 24 |
+
├── asph_nat_meth_test.tsv
|
| 25 |
+
├── scaled_re_bio_synth_training_data_cleaned_05_09_2023.tsv
|
| 26 |
+
├── scaled_re_nat_meth_test.tsv
|
| 27 |
+
├── scaled_rg_bio_synth_training_data_cleaned_05_09_2023.tsv
|
| 28 |
+
├── scaled_rg_nat_meth_test.tsv
|
| 29 |
+
├── scaling_exp_bio_synth_training_data_cleaned_05_09_2023.tsv
|
| 30 |
+
├── scaling_exp_nat_meth_test.tsv
|
| 31 |
+
```
|
| 32 |
+
- **`asph`**=asphericity, **`scaled_re`**=scaled R<sub>e</sub>, **`scaled_rg`**=scaled R<sub>g</sub>, **`scaling_exp`**=polymer scaling exponent
|
| 33 |
+
- **`<property>_bio_synth_training_data_cleaned_05_09_2023.tsv`** are ALBATROSS **training data** for the four properties, downloaded directly their GitHub
|
| 34 |
+
- **`<property>_nat_meth_test.tsv`** are ALBATROSS **testing data** for the four proeprties, downloaded directly from their GitHub
|
| 35 |
+
|
| 36 |
+
### Cleaning raw IDR data
|
| 37 |
+
`clean.py` cleans the raw training and testing data separately for each property. Any duplicates (in both train and test) are removed from train and kept in test. Finally, the four are combined into one file:
|
| 38 |
+
|
| 39 |
+
```
|
| 40 |
+
benchmarking/
|
| 41 |
+
└── idr_prediction/
|
| 42 |
+
└── processed_data/
|
| 43 |
+
├── all_albatross_seqs_and_properties.csv
|
| 44 |
+
```
|
| 45 |
+
|
| 46 |
+
- **`all_albatross_seqs_and_properties.csv`**: Columns = "Sequence","IDs","UniProt_IDs","UniProt_Names","Split","asph","scaled_re","scaled_rg","scaling_exp". All splits are either "Train" or "Test", indicating ALBATROSS model's usage of them
|
| 47 |
+
|
| 48 |
+
To perform cleaning, run
|
| 49 |
+
|
| 50 |
+
```
|
| 51 |
+
python clean.py
|
| 52 |
+
```
|
| 53 |
+
|
| 54 |
+
### Using config.py for clustering, splitting, training
|
| 55 |
+
|
| 56 |
+
This file has configurations for clustering, splitting, training.
|
| 57 |
+
|
| 58 |
+
```
|
| 59 |
+
# Clustering Parameters
|
| 60 |
+
CLUSTER = CustomParams(
|
| 61 |
+
# MMSeqs2 parameters: see GitHub or MMSeqs2 Wiki for guidance
|
| 62 |
+
MIN_SEQ_ID = 0.3, # % identity
|
| 63 |
+
C = 0.5, # % sequence length overlap
|
| 64 |
+
COV_MODE = 1, # cov-mode: 0 = bidirectional, 1 = target coverage, 2 = query coverage, 3 = target-in-query length coverage.
|
| 65 |
+
CLUSTER_MODE = 2,
|
| 66 |
+
# File paths
|
| 67 |
+
INPUT_PATH = 'processed_data/all_albatross_seqs_and_properties.csv',
|
| 68 |
+
PATH_TO_MMSEQS = '../../mmseqs' # path to where you installed MMSeqs2
|
| 69 |
+
)
|
| 70 |
+
|
| 71 |
+
# Split config
|
| 72 |
+
SPLIT = CustomParams(
|
| 73 |
+
IDR_DB_PATH = 'processed_data/all_albatross_seqs_and_properties.csv',
|
| 74 |
+
CLUSTER_OUTPUT_PATH = 'clustering/mmseqs_full_results.csv',
|
| 75 |
+
RANDOM_STATE_1 = 2, # random_state_1 = state for splitting all data into train & other
|
| 76 |
+
TEST_SIZE_1 = 0.21, # test size for data -> train/test split. e.g. 20 means 80% clusters in train, 20% clusters in other
|
| 77 |
+
RANDOM_STATE_2 = 6, # random_state_2 = state for splitting other from ^ into val and test
|
| 78 |
+
TEST_SIZE_2 = 0.50 # test size for train -> train/val split. e.g. 0.50 means 50% clusters in train, 50% clusters in test
|
| 79 |
+
|
| 80 |
+
)
|
| 81 |
+
|
| 82 |
+
# Which models to benchmark
|
| 83 |
+
TRAIN = CustomParams(
|
| 84 |
+
BENCHMARK_FUSONPLM = True,
|
| 85 |
+
FUSONPLM_CKPTS= "FusOn-pLM", # Dictionary: key = run name, values = epochs, or string "FusOn-pLM"
|
| 86 |
+
BENCHMARK_ESM = True,
|
| 87 |
+
|
| 88 |
+
# GPU configs
|
| 89 |
+
CUDA_VISIBLE_DEVICES="0",
|
| 90 |
+
|
| 91 |
+
# Overwriting configs
|
| 92 |
+
PERMISSION_TO_OVERWRITE_EMBEDDINGS = False, # if False, script will halt if it believes these embeddings have already been made.
|
| 93 |
+
PERMISSION_TO_OVERWRITE_MODELS = False # if False, script will halt if it believes these embeddings have already been made.
|
| 94 |
+
)
|
| 95 |
+
```
|
| 96 |
+
|
| 97 |
+
### Clustering
|
| 98 |
+
Clustering of all sequences in `all_albatross_seqs_and_properties.csv` is performed by `cluster.py`.
|
| 99 |
+
|
| 100 |
+
The clustering command entered by the script is:
|
| 101 |
+
```
|
| 102 |
+
mmseqs easy-cluster clustering/input.fasta clustering/raw_output/mmseqs clustering/raw_output --min-seq-id 0.3 -c 0.5 --cov-mode 1 --cluster-mode 2 --dbtype 1
|
| 103 |
+
```
|
| 104 |
+
The script will generate the following files:
|
| 105 |
+
```
|
| 106 |
+
benchmarking/
|
| 107 |
+
└── idr_prediction/
|
| 108 |
+
└── clustering/
|
| 109 |
+
├── input.fasta
|
| 110 |
+
├── mmseqs_full_results.csv
|
| 111 |
+
```
|
| 112 |
+
- **`clustering/input.fasta`**: the input file used by MMSeqs2 to cluster the fusion oncoprotein sequences. Headers are our assigned sequence IDs (can be found in the `IDs` column of `processed_data/all_albatross_seqs_and_properties.csv`.)
|
| 113 |
+
- **`clustering/mmseqs_full_results.csv`**: clustering results. Columns:
|
| 114 |
+
- `representative seq_id`: the seq_id of the sequence representing this cluster
|
| 115 |
+
- `member seq_id`: the seq_id of a member of the cluster
|
| 116 |
+
- `representative seq`: the amino acid sequence of the cluster representative (representative seq_id)
|
| 117 |
+
- `member seq`: the amino acid sequence of the cluster member
|
| 118 |
+
|
| 119 |
+
### Splitting
|
| 120 |
+
Cluster-based splitting is performed by `split.py`. Results are formatted as follows:
|
| 121 |
+
|
| 122 |
+
```
|
| 123 |
+
benchmarking/
|
| 124 |
+
└── idr_prediction/
|
| 125 |
+
└── splits/
|
| 126 |
+
└── asph/
|
| 127 |
+
├── test_df.csv
|
| 128 |
+
├── val_df.csv
|
| 129 |
+
├── train_df.csv
|
| 130 |
+
└── scaled_re/... # same format as splits/asph
|
| 131 |
+
└── scaled_rg/... # same format as splits/asph
|
| 132 |
+
└── scaling_exp/... # same format as splits/asph
|
| 133 |
+
├── test_cluster_split.csv
|
| 134 |
+
├── train_cluster_split.csv
|
| 135 |
+
├── val_cluster_split.csv
|
| 136 |
+
```
|
| 137 |
+
|
| 138 |
+
- **`<split>_cluster_split.csv`**: cluster information for the clusters in each split (train, val, test). Columns = "representative seq_id", "member seq_id", "representative seq", "member seq", "member length"
|
| 139 |
+
- 📁 **`asph/`**, **`scaled_re/`**, **`scaled_rg/`**, and **`scaling_exp/`** contain the train, val, and test sets for each property (`train_df.csv`, `val_df.csv`, and `test_df.csv`). The splits follow `<split>_cluster_split.csv`, but not every property has a measurement for each of these sequences. The train-val-test ratio still remains 80-10-10 for each property, despite the sequence losses.
|
| 140 |
+
|
| 141 |
+
### Training
|
| 142 |
+
The model is defined in `model.py` and `utils.py`. The `train.py` script trains FusOn-pLM-IDR and ESM-2-650M-IDR models *separately for each property* (asphericity, R<sub>e</sub>, R<sub>g</sub>, scaling exponent) with a hyperparameter screen, saves all results separated by property, and makes plots. `plot.py` can be used to regenerate the R<sup>2</sup> plots.
|
| 143 |
+
- All **results** are stored in `idr_prediction/results/<timestamp>`, where `timestamp` is a unique string encoding the date and time when you started training.
|
| 144 |
+
- All **raw outputs from models** are stored in `idr_prediction/trained_models/<embedding_path>`, where `embedding_path` represents the embeddings used to build the disorder predictor.
|
| 145 |
+
- All **embeddings** made for training will be stored in a new folder called `idr_prediction/embeddings/` with subfolders for each model. This allows you to use the same model multiple times without regenerating embeddings.
|
| 146 |
+
|
| 147 |
+
Below is the FusOn-pLM-Diso raw outputs folder, `trained_models/fuson_plm/best/`, and the results from the paper, `results/final/`...
|
| 148 |
+
|
| 149 |
+
The outputs are structured as follows:
|
| 150 |
+
|
| 151 |
+
```
|
| 152 |
+
benchmarking/
|
| 153 |
+
└── idr_prediction/
|
| 154 |
+
└── results/final/
|
| 155 |
+
└── r2_plots
|
| 156 |
+
└── asph/
|
| 157 |
+
├── esm2_t33_650M_UR50D_asph_R2.png
|
| 158 |
+
├── esm2_t33_650M_UR50D_asph_R2_source_data.csv
|
| 159 |
+
├── fuson_plm_asph_R2.png
|
| 160 |
+
├── fuson_plm_asph_R2_source_data.csv
|
| 161 |
+
└── scaled_re/ # same format as r2_plots/asph/...
|
| 162 |
+
└── scaled_rg/ # same format as r2_plots/asph/...
|
| 163 |
+
└── scaling_exp/ # same format as r2_plots/asph/...
|
| 164 |
+
├── asph_best_test_r2.csv
|
| 165 |
+
├── asph_hyperparam_screen_test_r2.csv
|
| 166 |
+
├── scaled_re_best_test_r2.csv
|
| 167 |
+
├── scaled_re_hyperparam_screen_test_r2.csv
|
| 168 |
+
├── scaled_rg_best_test_r2.csv
|
| 169 |
+
├── scaled_rg_hyperparam_screen_test_r2.csv
|
| 170 |
+
├── scaling_exp_best_test_r2.csv
|
| 171 |
+
├── scaling_exp_hyperparam_screen_test_r2.csv
|
| 172 |
+
└── trained_models/
|
| 173 |
+
└── asph/
|
| 174 |
+
└── fuson_plm/best/
|
| 175 |
+
└── lr0.0001_bs32/
|
| 176 |
+
├── asph_r2.csv
|
| 177 |
+
├── train_val_losses.csv
|
| 178 |
+
├── test_loss.csv
|
| 179 |
+
├── asph_test_predictions.csv
|
| 180 |
+
└── ... other hyperparameter folders with same format as lr0.001_bs32/
|
| 181 |
+
└── esm2_t33_650M_UR50D # same format as asph/fuson_plm/best/
|
| 182 |
+
└── scaled_re/ # same format as asph/
|
| 183 |
+
└── scaled_rg/ # same format as asph/
|
| 184 |
+
└── scaling_exp/ # same format as asph/
|
| 185 |
+
|
| 186 |
+
```
|
| 187 |
+
|
| 188 |
+
In both directories, results are organized by IDR property and by the type of embedding used to train FusOn-pLM-IDR.
|
| 189 |
+
|
| 190 |
+
In the 📁 `results/final` directory.
|
| 191 |
+
- 📁 **`r2_plots/<property>/`**: holds all R<sup>2</sup> plots and source data (the formatted data used to make the R<sup>2</sup> plots) for these properties.
|
| 192 |
+
- **`<property>_best_test_r2.csv`**: holds the R<sup>2</sup> values for the top-performing models of each embedding type (e.g. ESM-2-650M and a specific checkpoint of FusOn-pLM)
|
| 193 |
+
- **`<property>_hyperparam_screen_test_r2.csv`**: holds the R<sup>2</sup> values for all embedding types, for all screened hyperparaemters
|
| 194 |
+
|
| 195 |
+
In the 📁 `trained_models` directory:
|
| 196 |
+
- 📁 `<property>/`: holds all results for all trained models predicting this property
|
| 197 |
+
- 📁 `asph/fuson_plm/best/`: holds all FusOn-pLM-IDR results on asphericity prediction for each set of hyperparameters screened when embeddings are made from "fuson_plm/best" (FusOn-pLM model). For example, 📁 `lr0.0001_bs32/` holds results for learning rate of 0.001, batch size 32. If you were to retrain your own checkpoint of fuson_plm and run the IDR prediction benchmark, its results would be stored in a new subfolder of `trained_models/fuson_plm`.
|
| 198 |
+
- **`asph/fuson_plm/best/lr0.0001_bs32/asph_r2.csv`**: R<sup>2</sup> value for this set of hyperparameters with "fuson_plm/best" embeddings
|
| 199 |
+
- **`asph/fuson_plm/best/lr0.0001_bs32/asph_test_predictions.csv`**: true asphericity values of the test set proteins, alongside FusOn-pLM-IDR's predictions of them.
|
| 200 |
+
- **`asph/fuson_plm/best/lr0.0001_bs32/test_loss.csv`**: FusOn-pLM-IDR's asphericity test loss value
|
| 201 |
+
- **`asph/fuson_plm/best/lr0.0001_bs32/train_val_losses.csv`**: FusOn-pLM-IDR's tarining and validation loss over each epoch while training on asphericity data
|
| 202 |
+
|
| 203 |
+
To run the training script, enter:
|
| 204 |
+
```
|
| 205 |
+
nohup python train.py > train.out 2> train.err &
|
| 206 |
+
```
|
| 207 |
+
|
| 208 |
+
To run the plotting script, enter:
|
| 209 |
+
```
|
| 210 |
+
python plot.py
|
| 211 |
+
```
|
fuson_plm/benchmarking/idr_prediction/__init__.py
ADDED
|
File without changes
|
fuson_plm/benchmarking/idr_prediction/clean.py
ADDED
|
@@ -0,0 +1,289 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
import numpy as np
|
| 3 |
+
import os
|
| 4 |
+
from fuson_plm.utils.logging import open_logfile, log_update
|
| 5 |
+
from fuson_plm.utils.constants import DELIMITERS, VALID_AAS
|
| 6 |
+
from fuson_plm.utils.data_cleaning import check_columns_for_listlike, find_invalid_chars
|
| 7 |
+
from fuson_plm.benchmarking.idr_prediction.plot import plot_all_values_hist_grid, plot_all_train_val_test_values_hist_grid
|
| 8 |
+
|
| 9 |
+
def process_raw_albatross(df):
|
| 10 |
+
# return a version of the df with first column split, duplicates cleaned,columns checked for weird characters and invalids
|
| 11 |
+
|
| 12 |
+
# first, look at the splits
|
| 13 |
+
split_str = df['Split'].value_counts().reset_index().rename(columns={'index': 'Split','Split': 'count'})
|
| 14 |
+
tot_prots = sum(split_str['count'])
|
| 15 |
+
split_str['pcnt'] = round(100*split_str['count']/tot_prots,2)
|
| 16 |
+
split_str = split_str.to_string(index=False)
|
| 17 |
+
split_str = "\t\t" + split_str.replace("\n","\n\t\t")
|
| 18 |
+
log_update(f"\tTotal proteins: {tot_prots}\n\tSplits:\n{split_str}")
|
| 19 |
+
|
| 20 |
+
# format: IDR_19076_tr___A0A8M9PNM5___A0A8M9PNM5_DANRE
|
| 21 |
+
# or: synth_test_sequence0
|
| 22 |
+
df['temp'] = df['ID'].str.split("_")
|
| 23 |
+
df['ID'] = df['temp'].apply(lambda x: f"{x[0]}" if len(x)==1 else f"{x[0]}_{x[1]}" if len(x)<3 else f"{x[0]}_{x[1]}_{x[2]}")
|
| 24 |
+
# Not ever column has UniProt IDs and Names, so we have to allow np.nan if this info is missing.
|
| 25 |
+
df['UniProt_ID'] = df['temp'].apply(lambda x: x[5].strip() if len(x)>=5 else np.nan)
|
| 26 |
+
df['UniProt_Name'] = df['temp'].apply(lambda x: f"{x[8].strip()}_{x[9].strip()}" if len(x)>=8 else np.nan)
|
| 27 |
+
df = df.drop(columns=['temp'])
|
| 28 |
+
|
| 29 |
+
cols_to_check = list(df.columns)
|
| 30 |
+
cols_to_check.remove('Value') # don't check this one because it shouldn't be string
|
| 31 |
+
# Investigate the colimns we just created and make sure they don't have any invalid features.
|
| 32 |
+
# make sure value is float type
|
| 33 |
+
assert df['Value'].dtype == 'float64'
|
| 34 |
+
check_columns_for_listlike(df, cols_of_interest=cols_to_check, delimiters=DELIMITERS)
|
| 35 |
+
|
| 36 |
+
# Check for invalid AAs
|
| 37 |
+
df['invalid_chars'] = df['Sequence'].apply(lambda x: find_invalid_chars(x, VALID_AAS))
|
| 38 |
+
df[df['invalid_chars'].str.len()>0].sort_values(by='Sequence')
|
| 39 |
+
all_invalid_chars = set().union(*df['invalid_chars'])
|
| 40 |
+
log_update(f"\tchecking for invalid characters...\n\t\tset of all invalid characters discovered within train_df: {all_invalid_chars}")
|
| 41 |
+
|
| 42 |
+
# Assert no invalid AAs
|
| 43 |
+
assert (df['invalid_chars'].str.len()==0).all()
|
| 44 |
+
df = df.drop(columns=['invalid_chars'])
|
| 45 |
+
|
| 46 |
+
# Check for duplicates - if we find any, REMOVE them from train and keep them in test
|
| 47 |
+
duplicates = df[df.duplicated('Sequence')]['Sequence'].unique().tolist()
|
| 48 |
+
n_rows_with_duplicates = len(df[df['Sequence'].isin(duplicates)])
|
| 49 |
+
log_update(f"\t{len(duplicates)} duplicated sequences, corresponding to {n_rows_with_duplicates} rows")
|
| 50 |
+
|
| 51 |
+
# Look for distribution of duplicates WITHIN train, WITHIN test, and BETWEEN train and test
|
| 52 |
+
# Train only
|
| 53 |
+
duplicates = df.loc[
|
| 54 |
+
(df['Split']=='Train')
|
| 55 |
+
]
|
| 56 |
+
duplicates = duplicates[duplicates.duplicated('Sequence')]['Sequence'].unique().tolist()
|
| 57 |
+
n_rows_with_duplicates = len(df.loc[
|
| 58 |
+
(df['Sequence'].isin(duplicates)) &
|
| 59 |
+
(df['Split']=='Train')
|
| 60 |
+
])
|
| 61 |
+
log_update(f"\t\twithin TRAIN only: {len(duplicates)} duplicated sequences, corresponding to {n_rows_with_duplicates} Train rows")
|
| 62 |
+
|
| 63 |
+
# Test only
|
| 64 |
+
duplicates = df.loc[
|
| 65 |
+
(df['Split']=='Test')
|
| 66 |
+
]
|
| 67 |
+
duplicates = duplicates[duplicates.duplicated('Sequence')]['Sequence'].unique().tolist()
|
| 68 |
+
n_rows_with_duplicates = len(df.loc[
|
| 69 |
+
(df['Sequence'].isin(duplicates)) &
|
| 70 |
+
(df['Split']=='Test')
|
| 71 |
+
])
|
| 72 |
+
log_update(f"\t\twithin TEST only: {len(duplicates)} duplicated sequences, corresponding to {n_rows_with_duplicates} Test rows")
|
| 73 |
+
|
| 74 |
+
# Between train and test
|
| 75 |
+
duplicates_df = df.groupby('Sequence').agg({
|
| 76 |
+
'Split': lambda x: ','.join(set(x))
|
| 77 |
+
}).reset_index()
|
| 78 |
+
duplicates_df = duplicates_df.loc[duplicates_df['Split'].str.contains(',')].reset_index(drop=True)
|
| 79 |
+
duplicates = duplicates_df['Sequence'].unique().tolist()
|
| 80 |
+
n_rows_with_duplicates = len(df[df['Sequence'].isin(duplicates)])
|
| 81 |
+
log_update(f"\t\tduplicates in BOTH TRAIN AND TEST: {len(duplicates)} duplicated sequences, corresponding to {n_rows_with_duplicates} rows")
|
| 82 |
+
log_update(f"\t\tprinting portion of dataframe with train+test shared seqs:\n{duplicates_df.head(5)}")
|
| 83 |
+
|
| 84 |
+
log_update("\tGrouping by sequence, averaging values, and keeping any Train/Test duplicates in the Test set...")
|
| 85 |
+
df = df.replace(np.nan, '')
|
| 86 |
+
df = df.groupby('Sequence').agg(
|
| 87 |
+
Value=('Value', 'mean'),
|
| 88 |
+
Value_STD=('Value', 'std'),
|
| 89 |
+
IDs=('ID', lambda x: ','.join(x)),
|
| 90 |
+
UniProt_IDs=('UniProt_ID', lambda x: ','.join(x)),
|
| 91 |
+
UniProt_Names=('UniProt_Name', lambda x: ','.join(x)),
|
| 92 |
+
Split=('Split', lambda x: ','.join(x))
|
| 93 |
+
).reset_index()
|
| 94 |
+
for col in ['IDs','UniProt_IDs','UniProt_Names','Split']:
|
| 95 |
+
df[col] = df[col].apply(lambda x: [y for y in x.split(',')])
|
| 96 |
+
df[col] = df[col].apply(lambda x: ','.join(x))
|
| 97 |
+
df[col] = df[col].str.strip(',')
|
| 98 |
+
# make sure there are no commas left
|
| 99 |
+
assert len(df[df[col].str.contains(',,')])==0
|
| 100 |
+
# set Split to Test if test is in it
|
| 101 |
+
df['Split'] = df['Split'].apply(lambda x: 'Test' if 'Test' in x else 'Train')
|
| 102 |
+
|
| 103 |
+
# For anything that wasn't duplicated, Value_STD is nan
|
| 104 |
+
log_update("\tChecking coefficients of variation for averaged rows")
|
| 105 |
+
# calculate coefficient of variation, should be < 10
|
| 106 |
+
df['Value_CV'] = 100*df['Value_STD']/df['Value']
|
| 107 |
+
log_update(f"\t\tTotal rows with coefficient of variation (CV)\n\t\t\t<=10%: {len(df[df['Value_CV']<=10])}\n\t\t\t>10%: {len(df[df['Value_CV']>10])}\n\t\t\t>20%: {len(df[df['Value_CV']>20])}")
|
| 108 |
+
|
| 109 |
+
# Ensure there are no duplicates
|
| 110 |
+
assert len(df[df['Sequence'].duplicated()])==0
|
| 111 |
+
log_update(f"\tNo remaining duplicates: {len(df[df['Sequence'].duplicated()])==0}")
|
| 112 |
+
|
| 113 |
+
# Print the final distribution of train and test values
|
| 114 |
+
split_str = df['Split'].value_counts().reset_index().rename(columns={'index': 'Split','Split': 'count'})
|
| 115 |
+
tot_prots = sum(split_str['count'])
|
| 116 |
+
split_str['pcnt'] = round(100*split_str['count']/tot_prots,2)
|
| 117 |
+
split_str = split_str.to_string(index=False)
|
| 118 |
+
split_str = "\t\t" + split_str.replace("\n","\n\t\t")
|
| 119 |
+
log_update(f"\tTotal proteins: {tot_prots}\n\tSplits:\n{split_str}")
|
| 120 |
+
|
| 121 |
+
return df
|
| 122 |
+
|
| 123 |
+
def combine_albatross_seqs(asph, scaled_re, scaled_rg, scaling_exp):
|
| 124 |
+
log_update("\nCombining all four dataframes into one file of ALBATROSS sequences")
|
| 125 |
+
|
| 126 |
+
asph = asph[['Sequence','Value','IDs','UniProt_IDs','UniProt_Names','Split']].rename(columns={'Value':'asph'})
|
| 127 |
+
scaled_re = scaled_re[['Sequence','Value','IDs','UniProt_IDs','UniProt_Names','Split']].rename(columns={'Value':'scaled_re'})
|
| 128 |
+
scaled_rg = scaled_rg[['Sequence','Value','IDs','UniProt_IDs','UniProt_Names','Split']].rename(columns={'Value':'scaled_rg'})
|
| 129 |
+
scaling_exp = scaling_exp[['Sequence','Value','IDs','UniProt_IDs','UniProt_Names','Split']].rename(columns={'Value':'scaling_exp'})
|
| 130 |
+
|
| 131 |
+
combined = asph.merge(scaled_re, on='Sequence',how='outer',suffixes=('_asph', '_scaledre'))\
|
| 132 |
+
.merge(scaled_rg, on='Sequence',how='outer',suffixes=('_scaledre', '_scaledrg'))\
|
| 133 |
+
.merge(scaling_exp, on='Sequence',how='outer',suffixes=('_scaledrg', '_scalingexp')).fillna('')
|
| 134 |
+
|
| 135 |
+
# Make sure something that's in train for one is in train for all, and not test
|
| 136 |
+
combined['IDs'] = combined['IDs_asph']+','+combined['IDs_scaledre']+','+combined['IDs_scaledrg']+','+combined['IDs_scalingexp']
|
| 137 |
+
combined['UniProt_IDs'] = combined['UniProt_IDs_asph']+','+combined['UniProt_IDs_scaledre']+','+combined['UniProt_IDs_scaledrg']+','+combined['UniProt_IDs_scalingexp']
|
| 138 |
+
combined['UniProt_Names'] = combined['UniProt_Names_asph']+','+combined['UniProt_Names_scaledre']+','+combined['UniProt_Names_scaledrg']+','+combined['UniProt_Names_scalingexp']
|
| 139 |
+
combined['Split'] = combined['Split_asph']+','+combined['Split_scaledre']+','+combined['Split_scaledrg']+','+combined['Split_scalingexp']
|
| 140 |
+
|
| 141 |
+
# Make the lists clean
|
| 142 |
+
for col in ['IDs','UniProt_IDs','UniProt_Names','Split']:
|
| 143 |
+
combined[col] = combined[col].apply(lambda x: [y.strip() for y in x.split(',') if len(y)>0])
|
| 144 |
+
combined[col] = combined[col].apply(lambda x: ','.join(set(x)))
|
| 145 |
+
combined[col] = combined[col].str.strip(',')
|
| 146 |
+
# make sure there are no commas left
|
| 147 |
+
assert len(combined[combined[col].str.contains(',,')])==0
|
| 148 |
+
combined = combined[['Sequence','IDs','UniProt_IDs','UniProt_Names','Split','asph','scaled_re','scaled_rg','scaling_exp']] # drop unneeded merge relics
|
| 149 |
+
combined = combined.replace('',np.nan)
|
| 150 |
+
# Make sure there are no sequences where split is both train and test
|
| 151 |
+
log_update("\tChecking for any cases where a protein is Train for one IDR prediction task and Test for another (should NOT happen!)")
|
| 152 |
+
duplicates_df = combined.groupby('Sequence').agg({
|
| 153 |
+
'Split': lambda x: ','.join(set(x))
|
| 154 |
+
}).reset_index()
|
| 155 |
+
duplicates_df = duplicates_df.loc[duplicates_df['Split'].str.contains(',')].reset_index(drop=True)
|
| 156 |
+
duplicates = duplicates_df['Sequence'].unique().tolist()
|
| 157 |
+
n_rows_with_duplicates = len(combined[combined['Sequence'].isin(duplicates)])
|
| 158 |
+
log_update(f"\t\tsequences in BOTH TRAIN AND TEST: {len(duplicates)} sequences, corresponding to {n_rows_with_duplicates} rows")
|
| 159 |
+
if len(duplicates)>0:
|
| 160 |
+
log_update(f"\t\tprinting portion of assert len(combined[combined['asph'].notna()])==len(asph)dataframe with train+test shared seqs:\n{duplicates_df.head(5)}")
|
| 161 |
+
|
| 162 |
+
# Now, get rid of duplicates
|
| 163 |
+
combined = combined.drop_duplicates().reset_index(drop=True)
|
| 164 |
+
duplicates = combined[combined.duplicated('Sequence')]['Sequence'].unique().tolist()
|
| 165 |
+
log_update(f"\tDropped duplicates.\n\tTotal duplicate sequences: {len(duplicates)}\n\tTotal sequences: {len(combined)}")
|
| 166 |
+
assert len(duplicates)==0
|
| 167 |
+
|
| 168 |
+
# See how many columns have multiple entries for each
|
| 169 |
+
log_update(f"\tChecking how many sequences have multiple of the following: ID, UniProt ID, UniProt Name")
|
| 170 |
+
for col in ['IDs','UniProt_IDs','UniProt_Names','Split']:
|
| 171 |
+
n_multiple = len(combined.loc[(combined[col].notna()) & (combined[col].str.contains(','))])
|
| 172 |
+
log_update(f"\t\t{col}: {n_multiple}")
|
| 173 |
+
|
| 174 |
+
# See how many entries there are of each cproperty (should match length of original database)
|
| 175 |
+
assert len(combined[combined['asph'].notna()])==len(asph)
|
| 176 |
+
assert len(combined[combined['scaled_re'].notna()])==len(scaled_re)
|
| 177 |
+
assert len(combined[combined['scaled_rg'].notna()])==len(scaled_rg)
|
| 178 |
+
assert len(combined[combined['scaling_exp'].notna()])==len(scaling_exp)
|
| 179 |
+
log_update("\tSequences with values for each property:")
|
| 180 |
+
for property in ['asph','scaled_re','scaled_rg','scaling_exp']:
|
| 181 |
+
log_update(f"\t\t{property}: {len(combined[combined[property].notna()])}")
|
| 182 |
+
|
| 183 |
+
log_update(f"\nPreview of combined database with columns: {combined.columns}\n{combined.head(10)}")
|
| 184 |
+
return combined
|
| 185 |
+
|
| 186 |
+
def main():
|
| 187 |
+
with open_logfile("data_cleaning_log.txt"):
|
| 188 |
+
# Read in all of the raw data
|
| 189 |
+
raw_data_folder = 'raw_data'
|
| 190 |
+
dtype_dict = {0:str,1:str,2:float}
|
| 191 |
+
rename_dict = {0:'ID',1:'Sequence',2:'Value'}
|
| 192 |
+
|
| 193 |
+
# Read in the test data
|
| 194 |
+
asph_test = pd.read_csv(f"{raw_data_folder}/asph_nat_meth_test.tsv",sep=" ",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 195 |
+
scaled_re_test = pd.read_csv(f"{raw_data_folder}/scaled_re_nat_meth_test.tsv",sep="\t",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 196 |
+
scaled_rg_test = pd.read_csv(f"{raw_data_folder}/scaled_rg_nat_meth_test.tsv",sep="\t",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 197 |
+
scaling_exp_test = pd.read_csv(f"{raw_data_folder}/scaling_exp_nat_meth_test.tsv",sep=" ",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 198 |
+
|
| 199 |
+
# Read in the train data
|
| 200 |
+
asph_train = pd.read_csv(f"{raw_data_folder}/asph_bio_synth_training_data_cleaned_05_09_2023.tsv",sep=" ",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 201 |
+
scaled_re_train = pd.read_csv(f"{raw_data_folder}/scaled_re_bio_synth_training_data_cleaned_05_09_2023.tsv",sep="\t",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 202 |
+
scaled_rg_train = pd.read_csv(f"{raw_data_folder}/scaled_rg_bio_synth_training_data_cleaned_05_09_2023.tsv",sep="\t",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 203 |
+
scaling_exp_train = pd.read_csv(f"{raw_data_folder}/scaling_exp_bio_synth_training_data_cleaned_05_09_2023.tsv",sep=" ",dtype=dtype_dict,header=None).rename(columns=rename_dict)
|
| 204 |
+
|
| 205 |
+
# Concatenate - include columns for split
|
| 206 |
+
asph_test['Split'] = ['Test']*len(asph_test)
|
| 207 |
+
scaled_re_test['Split'] = ['Test']*len(scaled_re_test)
|
| 208 |
+
scaled_rg_test['Split'] = ['Test']*len(scaled_rg_test)
|
| 209 |
+
scaling_exp_test['Split'] = ['Test']*len(scaling_exp_test)
|
| 210 |
+
|
| 211 |
+
asph_train['Split'] = ['Train']*len(asph_train)
|
| 212 |
+
scaled_re_train['Split'] = ['Train']*len(scaled_re_train)
|
| 213 |
+
scaled_rg_train['Split'] = ['Train']*len(scaled_rg_train)
|
| 214 |
+
scaling_exp_train['Split'] = ['Train']*len(scaling_exp_train)
|
| 215 |
+
|
| 216 |
+
asph = pd.concat([asph_test, asph_train])
|
| 217 |
+
scaled_re = pd.concat([scaled_re_test, scaled_re_train])
|
| 218 |
+
scaled_rg = pd.concat([scaled_rg_test, scaled_rg_train])
|
| 219 |
+
scaling_exp = pd.concat([scaling_exp_test, scaling_exp_train])
|
| 220 |
+
|
| 221 |
+
log_update("Initial counts:")
|
| 222 |
+
log_update(f"\tAsphericity: total entries={len(asph)}, not nan entries={len(asph.loc[asph['Value'].notna()])}")
|
| 223 |
+
log_update(f"\tScaled re: total entries={len(scaled_re)}, not nan entries={len(scaled_re.loc[scaled_re['Value'].notna()])}")
|
| 224 |
+
log_update(f"\tScaled rg: total entries={len(scaled_rg)}, not nan entries={len(scaled_rg.loc[scaled_rg['Value'].notna()])}")
|
| 225 |
+
# change any scaled_rg rows with values less than 1 to np.nan, as done in the paper
|
| 226 |
+
scaled_rg = scaled_rg.loc[
|
| 227 |
+
scaled_rg['Value']>=1].reset_index(drop=True)
|
| 228 |
+
log_update(f"\t\tAfter dropping Rg values < 1: total entries={len(scaled_rg)}")
|
| 229 |
+
log_update(f"\tScaling exp: total entries={len(scaling_exp)}, not nan entries={len(scaling_exp.loc[scaling_exp['Value'].notna()])}")
|
| 230 |
+
|
| 231 |
+
# Process the raw data
|
| 232 |
+
log_update(f"Example raw download: asphericity\n{asph.head()}")
|
| 233 |
+
log_update(f"\nCleaning Asphericity")
|
| 234 |
+
asph = process_raw_albatross(asph)
|
| 235 |
+
log_update(f"\nProcessed data: asphericity\n{asph.head()}")
|
| 236 |
+
|
| 237 |
+
log_update(f"\nCleaning Scaled Re")
|
| 238 |
+
scaled_re = process_raw_albatross(scaled_re)
|
| 239 |
+
log_update(f"\nProcessed data: scaled re\n{scaled_re.head()}")
|
| 240 |
+
|
| 241 |
+
log_update(f"\nCleaning Scaled Rg")
|
| 242 |
+
scaled_rg = process_raw_albatross(scaled_rg)
|
| 243 |
+
log_update(f"\nProcessed data: scaled rg\n{scaled_rg.head()}")
|
| 244 |
+
|
| 245 |
+
log_update(f"\nCleaning Scaling Exp")
|
| 246 |
+
scaling_exp = process_raw_albatross(scaling_exp)
|
| 247 |
+
log_update(f"\nProcessed data: scaling exp\n{scaling_exp.head()}")
|
| 248 |
+
|
| 249 |
+
# Give some stats about each dataset
|
| 250 |
+
log_update("\nStats:")
|
| 251 |
+
log_update(f"# Asphericity sequences: {len(asph)}\n\tRange: {min(asph['Value']):.4f}-{max(asph['Value']):.4f}")
|
| 252 |
+
log_update(f"# Scaled Re sequences: {len(scaled_re)}\n\tRange: {min(scaled_re['Value']):.4f}-{max(scaled_re['Value']):.4f}")
|
| 253 |
+
log_update(f"# Scaled Rg sequences: {len(scaled_rg)}\n\tRange: {min(scaled_rg['Value']):.4f}-{max(scaled_rg['Value']):.4f}")
|
| 254 |
+
log_update(f"# Scaling Exponent sequences: {len(scaling_exp)}\n\tRange: {min(scaling_exp['Value']):.4f}-{max(scaling_exp['Value']):.4f}")
|
| 255 |
+
|
| 256 |
+
# Combine
|
| 257 |
+
combined = combine_albatross_seqs(asph, scaled_re, scaled_rg, scaling_exp)
|
| 258 |
+
|
| 259 |
+
# Save processed data
|
| 260 |
+
proc_folder = "processed_data"
|
| 261 |
+
os.makedirs(proc_folder,exist_ok=True)
|
| 262 |
+
combined.to_csv(f"{proc_folder}/all_albatross_seqs_and_properties.csv",index=False)
|
| 263 |
+
|
| 264 |
+
# Plot the data distribution and save it
|
| 265 |
+
values_dict = {
|
| 266 |
+
'Asphericity': asph['Value'].tolist(),
|
| 267 |
+
'End-to-End Distance (Re)': scaled_re['Value'].tolist(),
|
| 268 |
+
'Radius of Gyration (Rg)': scaled_rg['Value'].tolist(),
|
| 269 |
+
'Scaling Exponent': scaling_exp['Value'].tolist()
|
| 270 |
+
}
|
| 271 |
+
train_test_values_dict = {
|
| 272 |
+
'Asphericity': {
|
| 273 |
+
'train': asph[asph['Split']=='Train']['Value'].tolist(),
|
| 274 |
+
'test': asph[asph['Split']=='Test']['Value'].tolist()},
|
| 275 |
+
'End-to-End Distance (Re)': {
|
| 276 |
+
'train': scaled_re[scaled_re['Split']=='Train']['Value'].tolist(),
|
| 277 |
+
'test': scaled_re[scaled_re['Split']=='Test']['Value'].tolist()},
|
| 278 |
+
'Radius of Gyration (Rg)': {
|
| 279 |
+
'train': scaled_rg[scaled_rg['Split']=='Train']['Value'].tolist(),
|
| 280 |
+
'test': scaled_rg[scaled_rg['Split']=='Test']['Value'].tolist()},
|
| 281 |
+
'Scaling Exponent': {
|
| 282 |
+
'train': scaling_exp[scaling_exp['Split']=='Train']['Value'].tolist(),
|
| 283 |
+
'test': scaling_exp[scaling_exp['Split']=='Test']['Value'].tolist()},
|
| 284 |
+
}
|
| 285 |
+
plot_all_values_hist_grid(values_dict, save_path="processed_data/value_histograms.png")
|
| 286 |
+
plot_all_train_val_test_values_hist_grid(train_test_values_dict, save_path="processed_data/train_test_value_histograms.png")
|
| 287 |
+
|
| 288 |
+
if __name__ == "__main__":
|
| 289 |
+
main()
|
fuson_plm/benchmarking/idr_prediction/cluster.py
ADDED
|
@@ -0,0 +1,94 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from fuson_plm.utils.logging import open_logfile, log_update
|
| 2 |
+
from fuson_plm.benchmarking.idr_prediction.config import CLUSTER
|
| 3 |
+
from fuson_plm.utils.clustering import ensure_mmseqs_in_path, process_fasta, analyze_clustering_result, make_fasta, run_mmseqs_clustering, cluster_summary
|
| 4 |
+
import os
|
| 5 |
+
import pandas as pd
|
| 6 |
+
|
| 7 |
+
def main_old():
|
| 8 |
+
# Read all the input args
|
| 9 |
+
LOG_PATH = "clustering_log.txt"
|
| 10 |
+
INPUT_PATH = CLUSTER.INPUT_PATH
|
| 11 |
+
MIN_SEQ_ID = CLUSTER.MIN_SEQ_ID
|
| 12 |
+
C = CLUSTER.C
|
| 13 |
+
COV_MODE = CLUSTER.COV_MODE
|
| 14 |
+
CLUSTER_MODE = CLUSTER.CLUSTER_MODE
|
| 15 |
+
PATH_TO_MMSEQS = CLUSTER.PATH_TO_MMSEQS
|
| 16 |
+
|
| 17 |
+
with open_logfile(LOG_PATH):
|
| 18 |
+
log_update("Input params from config.py:")
|
| 19 |
+
CLUSTER.print_config(indent='\t')
|
| 20 |
+
# Make a subfolder for clustering results, and direct MMSeqs2 outputs here
|
| 21 |
+
os.makedirs("clustering",exist_ok=True)
|
| 22 |
+
output_dir = "clustering/raw_output"
|
| 23 |
+
|
| 24 |
+
# Make fasta of input file
|
| 25 |
+
sequences = pd.read_csv(INPUT_PATH)
|
| 26 |
+
# We only want to cluster the ones in the Train split from Albatross
|
| 27 |
+
sequences = sequences.loc[sequences['Split']=='Train'].reset_index(drop=True)
|
| 28 |
+
log_update(f"\nPreparing input data (albatross TRAIN only)...\n\tdataset size: {len(sequences)} sequences")
|
| 29 |
+
|
| 30 |
+
max_seqlen = max(sequences['Sequence'].str.len().tolist())
|
| 31 |
+
log_update(f"\tLongest sequence in dataset: {max_seqlen} AAs")
|
| 32 |
+
|
| 33 |
+
# Unfortunately, these IDs are NOT unique. Need to add tags to them
|
| 34 |
+
sequences['Unique_ID'] = [f"s{i+1}" for i in range(len(sequences))]
|
| 35 |
+
sequences['Unique_ID'] = sequences["IDs"].apply(lambda x: "_".join(x.split(','))) + "_" + sequences['Unique_ID']
|
| 36 |
+
log_update("Not all IDs from the database are unique. Created unique IDs by tagging on sequence #s")
|
| 37 |
+
log_update(f"\tExample: {sequences.iloc[0]['Unique_ID']}")
|
| 38 |
+
sequences = dict(zip(sequences['Unique_ID'],sequences['Sequence']))
|
| 39 |
+
fasta_path = make_fasta(sequences, "clustering/input.fasta")
|
| 40 |
+
log_update(f"\tMade fasta of input sequences, saved at {fasta_path}")
|
| 41 |
+
|
| 42 |
+
run_mmseqs_clustering(fasta_path, output_dir, min_seq_id=MIN_SEQ_ID, c=C, cov_mode=COV_MODE, cluster_mode=CLUSTER_MODE, path_to_mmseqs=PATH_TO_MMSEQS)
|
| 43 |
+
|
| 44 |
+
# Brief read to preview results
|
| 45 |
+
clusters = analyze_clustering_result('clustering/input.fasta', 'clustering/raw_output/mmseqs_cluster.tsv')
|
| 46 |
+
# Save clusters
|
| 47 |
+
clusters.to_csv('clustering/mmseqs_full_results.csv',index=False)
|
| 48 |
+
log_update("Processed and combined mmseqs output. Wrote comprehensive results to clustering/mmseqs_full_results.csv")
|
| 49 |
+
cluster_summary(clusters)
|
| 50 |
+
|
| 51 |
+
def main():
|
| 52 |
+
# Read all the input args
|
| 53 |
+
LOG_PATH = "clustering_log.txt"
|
| 54 |
+
INPUT_PATH = CLUSTER.INPUT_PATH
|
| 55 |
+
MIN_SEQ_ID = CLUSTER.MIN_SEQ_ID
|
| 56 |
+
C = CLUSTER.C
|
| 57 |
+
COV_MODE = CLUSTER.COV_MODE
|
| 58 |
+
CLUSTER_MODE = CLUSTER.CLUSTER_MODE
|
| 59 |
+
PATH_TO_MMSEQS = CLUSTER.PATH_TO_MMSEQS
|
| 60 |
+
|
| 61 |
+
with open_logfile(LOG_PATH):
|
| 62 |
+
log_update("Input params from config.py:")
|
| 63 |
+
CLUSTER.print_config(indent='\t')
|
| 64 |
+
# Make a subfolder for clustering results, and direct MMSeqs2 outputs here
|
| 65 |
+
os.makedirs("clustering",exist_ok=True)
|
| 66 |
+
output_dir = "clustering/raw_output"
|
| 67 |
+
|
| 68 |
+
# Make fasta of input file
|
| 69 |
+
sequences = pd.read_csv(INPUT_PATH)
|
| 70 |
+
log_update(f"\nPreparing input data (albatross train AND test sequences)...\n\tdataset size: {len(sequences)} sequences")
|
| 71 |
+
|
| 72 |
+
max_seqlen = max(sequences['Sequence'].str.len().tolist())
|
| 73 |
+
log_update(f"\tLongest sequence in dataset: {max_seqlen} AAs")
|
| 74 |
+
|
| 75 |
+
# Unfortunately, these IDs are NOT unique. Need to add tags to them
|
| 76 |
+
sequences['Unique_ID'] = [f"s{i+1}" for i in range(len(sequences))]
|
| 77 |
+
sequences['Unique_ID'] = sequences["IDs"].apply(lambda x: "_".join(x.split(','))) + "_" + sequences['Unique_ID']
|
| 78 |
+
log_update("Not all IDs from the database are unique. Created unique IDs by tagging on sequence #s")
|
| 79 |
+
log_update(f"\tExample: {sequences.iloc[0]['Unique_ID']}")
|
| 80 |
+
sequences = dict(zip(sequences['Unique_ID'],sequences['Sequence']))
|
| 81 |
+
fasta_path = make_fasta(sequences, "clustering/input.fasta")
|
| 82 |
+
log_update(f"\tMade fasta of input sequences, saved at {fasta_path}")
|
| 83 |
+
|
| 84 |
+
run_mmseqs_clustering(fasta_path, output_dir, min_seq_id=MIN_SEQ_ID, c=C, cov_mode=COV_MODE, cluster_mode=CLUSTER_MODE, path_to_mmseqs=PATH_TO_MMSEQS)
|
| 85 |
+
|
| 86 |
+
# Brief read to preview results
|
| 87 |
+
clusters = analyze_clustering_result('clustering/input.fasta', 'clustering/raw_output/mmseqs_cluster.tsv')
|
| 88 |
+
# Save clusters
|
| 89 |
+
clusters.to_csv('clustering/mmseqs_full_results.csv',index=False)
|
| 90 |
+
log_update("Processed and combined mmseqs output. Wrote comprehensive results to clustering/mmseqs_full_results.csv")
|
| 91 |
+
cluster_summary(clusters)
|
| 92 |
+
|
| 93 |
+
if __name__ == "__main__":
|
| 94 |
+
main()
|
fuson_plm/benchmarking/idr_prediction/clustering/input.fasta
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6445f3d52da17cd0843cf7ea8d49a38a3f122c4c852aec512e889a855e5aa373
|
| 3 |
+
size 7113231
|
fuson_plm/benchmarking/idr_prediction/clustering/mmseqs_full_results.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:fab007462284f7b9a3057d95aad0ab8257824b92ff9403b10443d790baafe5cd
|
| 3 |
+
size 16308334
|
fuson_plm/benchmarking/idr_prediction/config.py
ADDED
|
@@ -0,0 +1,41 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from fuson_plm.utils.logging import CustomParams
|
| 2 |
+
|
| 3 |
+
# Clustering Parameters
|
| 4 |
+
# Need to be stacked, because there are 4 properties
|
| 5 |
+
CLUSTER = CustomParams(
|
| 6 |
+
# MMSeqs2 parameters: see GitHub or MMSeqs2 Wiki for guidance
|
| 7 |
+
MIN_SEQ_ID = 0.3, # % identity
|
| 8 |
+
C = 0.5, # % sequence length overlap
|
| 9 |
+
COV_MODE = 1, # cov-mode: 0 = bidirectional, 1 = target coverage, 2 = query coverage, 3 = target-in-query length coverage.
|
| 10 |
+
CLUSTER_MODE = 2,
|
| 11 |
+
# File paths
|
| 12 |
+
INPUT_PATH = 'processed_data/all_albatross_seqs_and_properties.csv',
|
| 13 |
+
PATH_TO_MMSEQS = '../../mmseqs' # path to where you installed MMSeqs2
|
| 14 |
+
)
|
| 15 |
+
|
| 16 |
+
# Here, we'll be splitting the train set into train and val. we aren't touching test
|
| 17 |
+
SPLIT = CustomParams(
|
| 18 |
+
IDR_DB_PATH = 'processed_data/all_albatross_seqs_and_properties.csv',
|
| 19 |
+
CLUSTER_OUTPUT_PATH = 'clustering/mmseqs_full_results.csv',
|
| 20 |
+
#RANDOM_STATE = 7, # random_state_1 = state for splitting all data into train & test
|
| 21 |
+
#VAL_SIZE = 0.10, # val size for data -> train/val split. e.g. 20 means 80% clusters in train, 20% clusters in val
|
| 22 |
+
RANDOM_STATE_1 = 2, # random_state_1 = state for splitting all data into train & other
|
| 23 |
+
TEST_SIZE_1 = 0.21, # test size for data -> train/test split. e.g. 20 means 80% clusters in train, 20% clusters in other
|
| 24 |
+
RANDOM_STATE_2 = 6, # random_state_2 = state for splitting other from ^ into val and test
|
| 25 |
+
TEST_SIZE_2 = 0.50 # test size for train -> train/val split. e.g. 0.50 means 50% clusters in train, 50% clusters in test
|
| 26 |
+
|
| 27 |
+
)
|
| 28 |
+
|
| 29 |
+
# Which models to benchmark
|
| 30 |
+
TRAIN = CustomParams(
|
| 31 |
+
BENCHMARK_FUSONPLM = True,
|
| 32 |
+
FUSONPLM_CKPTS= "FusOn-pLM", # Dictionary: key = run name, values = epochs, or string "FusOn-pLM"
|
| 33 |
+
BENCHMARK_ESM = True,
|
| 34 |
+
|
| 35 |
+
# GPU configs
|
| 36 |
+
CUDA_VISIBLE_DEVICES="0",
|
| 37 |
+
|
| 38 |
+
# Overwriting configs
|
| 39 |
+
PERMISSION_TO_OVERWRITE_EMBEDDINGS = False, # if False, script will halt if it believes these embeddings have already been made.
|
| 40 |
+
PERMISSION_TO_OVERWRITE_MODELS = False # if False, script will halt if it believes these embeddings have already been made.
|
| 41 |
+
)
|
fuson_plm/benchmarking/idr_prediction/model.py
ADDED
|
@@ -0,0 +1,154 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pytorch_lightning as pl
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
import torch.nn.functional as F
|
| 4 |
+
from torch.utils.data import DataLoader
|
| 5 |
+
import pandas as pd
|
| 6 |
+
from tqdm import tqdm
|
| 7 |
+
import pickle
|
| 8 |
+
import torch
|
| 9 |
+
import esm
|
| 10 |
+
import numpy as np
|
| 11 |
+
import matplotlib.pyplot as plt
|
| 12 |
+
import random
|
| 13 |
+
import io
|
| 14 |
+
|
| 15 |
+
from transformers import EsmModel, EsmTokenizer, EsmConfig, AutoTokenizer
|
| 16 |
+
from sklearn.metrics import roc_auc_score
|
| 17 |
+
|
| 18 |
+
#one-hot MLP model (input 1280 (esm-2))
|
| 19 |
+
class ProteinMLPOneHot(pl.LightningModule):
|
| 20 |
+
def __init__(self):
|
| 21 |
+
super().__init__()
|
| 22 |
+
self.network = nn.Sequential(
|
| 23 |
+
nn.Linear(20, 8),
|
| 24 |
+
nn.ReLU(),
|
| 25 |
+
nn.LayerNorm(8),
|
| 26 |
+
nn.Dropout(0.2),
|
| 27 |
+
nn.Linear(8, 4),
|
| 28 |
+
nn.ReLU(),
|
| 29 |
+
nn.LayerNorm(4),
|
| 30 |
+
nn.Dropout(0.2),
|
| 31 |
+
nn.Linear(4, 1)
|
| 32 |
+
)
|
| 33 |
+
|
| 34 |
+
def forward(self, x):
|
| 35 |
+
x = self.network(x)
|
| 36 |
+
return x #pass x through linear layers with activation functions, dropout, and layernorm
|
| 37 |
+
|
| 38 |
+
def training_step(self, batch, batch_idx):
|
| 39 |
+
x, y = batch['Protein Input'], batch['Dimension'].float() #get batch
|
| 40 |
+
y_hat = self(x).squeeze(-1) #get prediction from batch
|
| 41 |
+
loss = F.mse_loss(y_hat, y) #calc loss from prediction and dimension of each
|
| 42 |
+
self.log('train_loss', loss, on_epoch=True, prog_bar=True, logger=True)
|
| 43 |
+
return loss
|
| 44 |
+
|
| 45 |
+
def validation_step(self, batch, batch_idx):
|
| 46 |
+
x, y = batch['Protein Input'], batch['Dimension'].float()
|
| 47 |
+
y_hat = self(x).squeeze(-1)
|
| 48 |
+
val_loss = F.mse_loss(y_hat, y)
|
| 49 |
+
self.log('val_loss', val_loss, on_epoch=True, prog_bar=True, logger=True)
|
| 50 |
+
return val_loss
|
| 51 |
+
|
| 52 |
+
def test_step(self, batch, batch_idx):
|
| 53 |
+
x, y = batch['Protein Input'], batch['Dimension'].float()
|
| 54 |
+
y_hat = self(x).squeeze(-1)
|
| 55 |
+
test_loss = F.mse_loss(y_hat, y)
|
| 56 |
+
self.log('test_loss', test_loss, on_epoch=True, prog_bar=True, logger=True)
|
| 57 |
+
return test_loss
|
| 58 |
+
|
| 59 |
+
def configure_optimizers(self):
|
| 60 |
+
optimizer = torch.optim.Adam(self.parameters(), lr=0.0003)
|
| 61 |
+
return optimizer
|
| 62 |
+
|
| 63 |
+
# def on_train_epoch_end(self):
|
| 64 |
+
# train_loss = self.trainer.callback_metrics['train_loss']
|
| 65 |
+
# print(f"Epoch {self.current_epoch + 1} - Training Loss: {train_loss:.4f}")
|
| 66 |
+
# wandb.log({'train_loss': train_loss, 'epoch': self.current_epoch + 1})
|
| 67 |
+
|
| 68 |
+
# def on_validation_epoch_end(self):
|
| 69 |
+
# val_loss = self.trainer.callback_metrics['val_loss']
|
| 70 |
+
# print(f"Epoch {self.current_epoch + 1} - Validation Loss: {val_loss:.4f}")
|
| 71 |
+
# wandb.log({'val_loss': val_loss, 'epoch': self.current_epoch + 1})
|
| 72 |
+
|
| 73 |
+
# def on_test_epoch_end(self):
|
| 74 |
+
# test_loss = self.trainer.callback_metrics['test_loss']
|
| 75 |
+
# print(f"Test Loss: {test_loss:.4f}")
|
| 76 |
+
# wandb.log({'test_loss': test_loss})
|
| 77 |
+
|
| 78 |
+
#regular MLP model (input 1280 (esm-2))
|
| 79 |
+
class ProteinMLPESM(pl.LightningModule):
|
| 80 |
+
def __init__(self):
|
| 81 |
+
super().__init__()
|
| 82 |
+
self.network = nn.Sequential(
|
| 83 |
+
nn.Linear(1280, 640),
|
| 84 |
+
nn.ReLU(),
|
| 85 |
+
nn.LayerNorm(640),
|
| 86 |
+
nn.Dropout(0.2),
|
| 87 |
+
nn.Linear(640, 320),
|
| 88 |
+
nn.ReLU(),
|
| 89 |
+
nn.LayerNorm(320),
|
| 90 |
+
nn.Dropout(0.2),
|
| 91 |
+
nn.Linear(320, 1)
|
| 92 |
+
)
|
| 93 |
+
|
| 94 |
+
def forward(self, x):
|
| 95 |
+
x = self.network(x)
|
| 96 |
+
return x #pass x through linear layers with activation functions, dropout, and layernorm
|
| 97 |
+
|
| 98 |
+
def training_step(self, batch, batch_idx):
|
| 99 |
+
x, y = batch['Protein Input'], batch['Dimension'].float() #get batch
|
| 100 |
+
y_hat = self(x).squeeze(-1) #get prediction from batch
|
| 101 |
+
loss = F.mse_loss(y_hat, y) #calc loss from prediction and dimension of each
|
| 102 |
+
self.log('train_loss', loss, on_epoch=True, prog_bar=True, logger=True)
|
| 103 |
+
return loss
|
| 104 |
+
|
| 105 |
+
def validation_step(self, batch, batch_idx):
|
| 106 |
+
x, y = batch['Protein Input'], batch['Dimension'].float()
|
| 107 |
+
y_hat = self(x).squeeze(-1)
|
| 108 |
+
val_loss = F.mse_loss(y_hat, y)
|
| 109 |
+
self.log('val_loss', val_loss, on_epoch=True, prog_bar=True, logger=True)
|
| 110 |
+
return val_loss
|
| 111 |
+
|
| 112 |
+
def test_step(self, batch, batch_idx):
|
| 113 |
+
x, y = batch['Protein Input'], batch['Dimension'].float()
|
| 114 |
+
y_hat = self(x).squeeze(-1)
|
| 115 |
+
test_loss = F.mse_loss(y_hat, y)
|
| 116 |
+
self.log('test_loss', test_loss, on_epoch=True, prog_bar=True, logger=True)
|
| 117 |
+
return test_loss
|
| 118 |
+
|
| 119 |
+
def configure_optimizers(self):
|
| 120 |
+
optimizer = torch.optim.Adam(self.parameters(), lr=0.0003)
|
| 121 |
+
return optimizer
|
| 122 |
+
|
| 123 |
+
# def on_train_epoch_end(self):
|
| 124 |
+
# train_loss = self.trainer.callback_metrics['train_loss']
|
| 125 |
+
# print(f"Epoch {self.current_epoch + 1} - Training Loss: {train_loss:.4f}")
|
| 126 |
+
# wandb.log({'train_loss': train_loss, 'epoch': self.current_epoch + 1})
|
| 127 |
+
|
| 128 |
+
# def on_validation_epoch_end(self):
|
| 129 |
+
# val_loss = self.trainer.callback_metrics['val_loss']
|
| 130 |
+
# print(f"Epoch {self.current_epoch + 1} - Validation Loss: {val_loss:.4f}")
|
| 131 |
+
# wandb.log({'val_loss': val_loss, 'epoch': self.current_epoch + 1})
|
| 132 |
+
|
| 133 |
+
# def on_test_epoch_end(self):
|
| 134 |
+
# test_loss = self.trainer.callback_metrics['test_loss']
|
| 135 |
+
# print(f"Test Loss: {test_loss:.4f}")
|
| 136 |
+
# wandb.log({'test_loss': test_loss})
|
| 137 |
+
|
| 138 |
+
|
| 139 |
+
class LossTrackerCallback(pl.Callback):
|
| 140 |
+
def __init__(self):
|
| 141 |
+
self.train_losses = []
|
| 142 |
+
self.val_losses = []
|
| 143 |
+
|
| 144 |
+
def on_train_epoch_end(self, trainer, pl_module):
|
| 145 |
+
# Access the most recent training loss from the logger
|
| 146 |
+
train_loss = trainer.callback_metrics.get('train_loss')
|
| 147 |
+
if train_loss:
|
| 148 |
+
self.train_losses.append(train_loss.item())
|
| 149 |
+
|
| 150 |
+
def on_validation_epoch_end(self, trainer, pl_module):
|
| 151 |
+
# Access the most recent validation loss from the logger
|
| 152 |
+
val_loss = trainer.callback_metrics.get('val_loss')
|
| 153 |
+
if val_loss:
|
| 154 |
+
self.val_losses.append(val_loss.item())
|
fuson_plm/benchmarking/idr_prediction/plot.py
ADDED
|
@@ -0,0 +1,204 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import matplotlib.pyplot as plt
|
| 2 |
+
import seaborn as sns
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import numpy as np
|
| 5 |
+
import os
|
| 6 |
+
from sklearn.metrics import r2_score
|
| 7 |
+
import matplotlib.colors as mcolors
|
| 8 |
+
from fuson_plm.utils.visualizing import set_font
|
| 9 |
+
|
| 10 |
+
global default_cmap_dict
|
| 11 |
+
default_cmap_dict = {
|
| 12 |
+
'Asphericity': '#785EF0',
|
| 13 |
+
'End-to-End Distance (Re)': '#DC267F',
|
| 14 |
+
'Radius of Gyration (Rg)': '#FE6100',
|
| 15 |
+
'Scaling Exponent': '#FFB000'
|
| 16 |
+
}
|
| 17 |
+
|
| 18 |
+
# Method for lengthening the model name
|
| 19 |
+
def lengthen_model_name(model_name, model_epoch):
|
| 20 |
+
if 'esm' in model_name:
|
| 21 |
+
return model_name
|
| 22 |
+
|
| 23 |
+
return f'{model_name}_e{model_epoch}'
|
| 24 |
+
|
| 25 |
+
def plot_train_val_test_values_hist(train_values_list, val_values_list, test_values_list, dataset_name="Data", color="black", save_path=None, ax=None):
|
| 26 |
+
"""
|
| 27 |
+
Plot Histogram to show the ranges of values
|
| 28 |
+
"""
|
| 29 |
+
set_font()
|
| 30 |
+
if ax is None:
|
| 31 |
+
fig, ax = plt.subplots(1, 1, figsize=(6, 4), dpi=300)
|
| 32 |
+
|
| 33 |
+
total_seqs = len(train_values_list)+len(val_values_list)+len(test_values_list)
|
| 34 |
+
ax.hist(train_values_list, color=color, alpha=0.7,label=f"train (n={len(train_values_list)})")
|
| 35 |
+
if not(test_values_list is None):
|
| 36 |
+
ax.hist(test_values_list, color='black',alpha=0.7,label=f"test (n={len(test_values_list)})")
|
| 37 |
+
if not(val_values_list is None):
|
| 38 |
+
ax.hist(val_values_list, color='grey',alpha=0.7,label=f"val (n={len(val_values_list)})")
|
| 39 |
+
ax.grid(True)
|
| 40 |
+
ax.set_axisbelow(True)
|
| 41 |
+
ax.set_title(f'{dataset_name} Distribution (n={total_seqs})')
|
| 42 |
+
ax.set_xlabel(dataset_name)
|
| 43 |
+
ax.legend()
|
| 44 |
+
plt.tight_layout()
|
| 45 |
+
|
| 46 |
+
if save_path is not None:
|
| 47 |
+
plt.savefig(save_path)
|
| 48 |
+
|
| 49 |
+
def plot_values_hist(values_list, dataset_name="Data", color="black", save_path=None, ax=None):
|
| 50 |
+
"""
|
| 51 |
+
Plot Histogram to show the ranges of values
|
| 52 |
+
"""
|
| 53 |
+
set_font()
|
| 54 |
+
if ax is None:
|
| 55 |
+
fig, ax = plt.subplots(1, 1, figsize=(6, 4), dpi=300)
|
| 56 |
+
|
| 57 |
+
ax.hist(values_list, color=color)
|
| 58 |
+
ax.grid(True)
|
| 59 |
+
ax.set_axisbelow(True)
|
| 60 |
+
ax.set_title(f'{dataset_name} Distribution')
|
| 61 |
+
ax.set_xlabel(dataset_name)
|
| 62 |
+
plt.tight_layout()
|
| 63 |
+
|
| 64 |
+
if save_path is not None:
|
| 65 |
+
plt.savefig(save_path)
|
| 66 |
+
|
| 67 |
+
def plot_all_values_hist_grid(values_dict, cmap_dict=default_cmap_dict, save_path="processed_data/value_histograms.png"):
|
| 68 |
+
"""
|
| 69 |
+
Args:
|
| 70 |
+
values_dict: dictionary where keys are dataset names and values are value lists
|
| 71 |
+
cmap_dict: dictioanry where keys are dataset names (same as in values dict) and values are value lists
|
| 72 |
+
"""
|
| 73 |
+
|
| 74 |
+
fig, axes = plt.subplots(2, 2, figsize=(12, 8), dpi=300)
|
| 75 |
+
axes = axes.flatten()
|
| 76 |
+
|
| 77 |
+
for i, (dataset_name, values_list) in enumerate(values_dict.items()):
|
| 78 |
+
ax = axes[i]
|
| 79 |
+
plot_values_hist(values_list, dataset_name=dataset_name, color=cmap_dict[dataset_name], ax=ax)
|
| 80 |
+
|
| 81 |
+
fig.set_tight_layout(True)
|
| 82 |
+
fig.savefig(save_path)
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
def plot_all_train_val_test_values_hist_grid(values_dict, cmap_dict=default_cmap_dict, save_path="processed_data/value_histograms.png"):
|
| 86 |
+
"""
|
| 87 |
+
Args:
|
| 88 |
+
values_dict: dictionary where keys are dataset names and values are another dict: {'train': train_values_list, 'test': test_values_list}
|
| 89 |
+
cmap_dict: dictioanry where keys are dataset names (same as in values dict) and values are value lists
|
| 90 |
+
"""
|
| 91 |
+
|
| 92 |
+
fig, axes = plt.subplots(2, 2, figsize=(12, 8), dpi=300)
|
| 93 |
+
axes = axes.flatten()
|
| 94 |
+
|
| 95 |
+
for i, (dataset_name, train_val_test_dict) in enumerate(values_dict.items()):
|
| 96 |
+
ax = axes[i]
|
| 97 |
+
train_values_list = train_val_test_dict['train']
|
| 98 |
+
test_values_list, val_values_list = None, None
|
| 99 |
+
if 'test' in train_val_test_dict:
|
| 100 |
+
test_values_list = train_val_test_dict['test']
|
| 101 |
+
if 'val' in train_val_test_dict:
|
| 102 |
+
val_values_list = train_val_test_dict['val']
|
| 103 |
+
plot_train_val_test_values_hist(train_values_list, val_values_list, test_values_list, dataset_name=dataset_name, color=cmap_dict[dataset_name], ax=ax)
|
| 104 |
+
|
| 105 |
+
fig.set_tight_layout(True)
|
| 106 |
+
fig.savefig(save_path)
|
| 107 |
+
|
| 108 |
+
#only need to change labels at bottom depending on what embeddings+dimension is being looked at
|
| 109 |
+
def plot_r2(model_type, idr_property, test_preds, save_path):
|
| 110 |
+
set_font()
|
| 111 |
+
|
| 112 |
+
# prepare ylabels from idr_property
|
| 113 |
+
ylabel_dict = {'asph': 'Asphericity',
|
| 114 |
+
'scaled_re': 'End-to-End Radius, $R_e$',
|
| 115 |
+
'scaled_rg': 'Radius of Gyration, $R_g$',
|
| 116 |
+
'scaling_exp': 'Polymer Scaling Exponent'}
|
| 117 |
+
y_unitlabel_dict = {'asph': 'Asphericity',
|
| 118 |
+
'scaled_re': '$R_e$ (Å)',
|
| 119 |
+
'scaled_rg': '$R_g$ (Å)',
|
| 120 |
+
'scaling_exp': 'Exponent'
|
| 121 |
+
|
| 122 |
+
}
|
| 123 |
+
y_label = ylabel_dict[idr_property]
|
| 124 |
+
y_unitlabel = y_unitlabel_dict[idr_property]
|
| 125 |
+
|
| 126 |
+
# get true values and predictions
|
| 127 |
+
true_values = test_preds['true_values'].tolist()
|
| 128 |
+
predictions = test_preds['predictions'].tolist()
|
| 129 |
+
|
| 130 |
+
# save this source data, including the IDs of the sequences
|
| 131 |
+
test_df = pd.read_csv(f"splits/{idr_property}/test_df.csv")
|
| 132 |
+
processed_data = pd.read_csv("processed_data/all_albatross_seqs_and_properties.csv")
|
| 133 |
+
seq_id_dict = dict(zip(processed_data['Sequence'],processed_data['IDs']))
|
| 134 |
+
test_df['IDs'] = test_df['Sequence'].map(seq_id_dict)
|
| 135 |
+
test_df_with_preds = test_preds[['true_values','predictions']]
|
| 136 |
+
test_df_with_preds['IDs'] = test_df['IDs']
|
| 137 |
+
print("number of sequences with no ID: ", len(test_df_with_preds.loc[test_df_with_preds['IDs'].isna()]))
|
| 138 |
+
test_df_with_preds.to_csv(save_path.replace(".png","_source_data.csv"),index=False)
|
| 139 |
+
|
| 140 |
+
r2 = r2_score(true_values, predictions)
|
| 141 |
+
|
| 142 |
+
# Plotting
|
| 143 |
+
plt.figure(figsize=(10, 8))
|
| 144 |
+
plt.scatter(true_values, predictions, alpha=0.5, label='Predictions')
|
| 145 |
+
plt.plot([min(true_values), max(true_values)], [min(true_values), max(true_values)], 'r--', label='Ideal Fit')
|
| 146 |
+
plt.text(0.65, 0.35, f"$R^2$ = {r2:.2f}", transform=plt.gca().transAxes, fontsize=44)
|
| 147 |
+
# Adjusting font sizes and setting font properties
|
| 148 |
+
plt.xlabel(f'True {y_unitlabel}',size=44)
|
| 149 |
+
plt.ylabel(f'Predicted {y_unitlabel}',size=44)
|
| 150 |
+
plt.title(f"{y_label}",size=50) #: {model_type}\n($R^2$={r2:.2f})",size=44)
|
| 151 |
+
|
| 152 |
+
# Create legend and set font properties
|
| 153 |
+
legend = plt.legend(fontsize=32)
|
| 154 |
+
for text in legend.get_texts():
|
| 155 |
+
text.set_fontsize(32)
|
| 156 |
+
|
| 157 |
+
# Adjust marker size in the legend
|
| 158 |
+
for handle in legend.legendHandles:
|
| 159 |
+
handle._sizes = [100]
|
| 160 |
+
|
| 161 |
+
# Enable grid
|
| 162 |
+
plt.grid(True)
|
| 163 |
+
|
| 164 |
+
# Adjusting tick labels font size
|
| 165 |
+
plt.xticks(fontsize=36)
|
| 166 |
+
plt.yticks(fontsize=36)
|
| 167 |
+
|
| 168 |
+
# Setting font properties for tick labels (another way to adjust them individually)
|
| 169 |
+
for label in plt.gca().get_xticklabels():
|
| 170 |
+
label.set_fontsize(32)
|
| 171 |
+
|
| 172 |
+
for label in plt.gca().get_yticklabels():
|
| 173 |
+
label.set_fontsize(32)
|
| 174 |
+
|
| 175 |
+
plt.tight_layout()
|
| 176 |
+
plt.savefig(save_path, dpi=300, bbox_inches='tight')
|
| 177 |
+
plt.show()
|
| 178 |
+
|
| 179 |
+
def plot_all_r2(output_dir, idr_properties):
|
| 180 |
+
for idr_property in idr_properties:
|
| 181 |
+
# make the R^2 Plots for the BEST one
|
| 182 |
+
best_results = pd.read_csv(f"{output_dir}/{idr_property}_best_test_r2.csv")
|
| 183 |
+
model_type_to_path_dict = dict(zip(best_results['model_type'],best_results['path_to_model']))
|
| 184 |
+
for model_type, path_to_model in model_type_to_path_dict.items():
|
| 185 |
+
model_preds_folder = path_to_model.split('/best-checkpoint.ckpt')[0]
|
| 186 |
+
test_preds = pd.read_csv(f"{model_preds_folder}/{idr_property}_test_predictions.csv")
|
| 187 |
+
|
| 188 |
+
# make paths for R^2 plots
|
| 189 |
+
if not os.path.exists(f"{output_dir}/r2_plots"):
|
| 190 |
+
os.makedirs(f"{output_dir}/r2_plots")
|
| 191 |
+
os.makedirs(f"{output_dir}/r2_plots/{idr_property}", exist_ok=True)
|
| 192 |
+
|
| 193 |
+
model_type_dict = {
|
| 194 |
+
'fuson_plm': 'FusOn-pLM',
|
| 195 |
+
'esm2_t33_650M_UR50D': 'ESM-2'
|
| 196 |
+
}
|
| 197 |
+
r2_save_path = f"{output_dir}/r2_plots/{idr_property}/{model_type}_{idr_property}_R2.png"
|
| 198 |
+
plot_r2(model_type_dict[model_type], idr_property, test_preds, r2_save_path)
|
| 199 |
+
|
| 200 |
+
def main():
|
| 201 |
+
plot_all_r2("results/final", ["asph","scaled_re","scaled_rg","scaling_exp"])
|
| 202 |
+
|
| 203 |
+
if __name__ == '__main__':
|
| 204 |
+
main()
|
fuson_plm/benchmarking/idr_prediction/processed_data/all_albatross_seqs_and_properties.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:9cb327d8990d40b566a399f3e075c6d5ccc73ca4203bc9dd9a94c00675e4dda5
|
| 3 |
+
size 9552773
|
fuson_plm/benchmarking/idr_prediction/processed_data/train_test_value_histograms.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/processed_data/value_histograms.png
ADDED
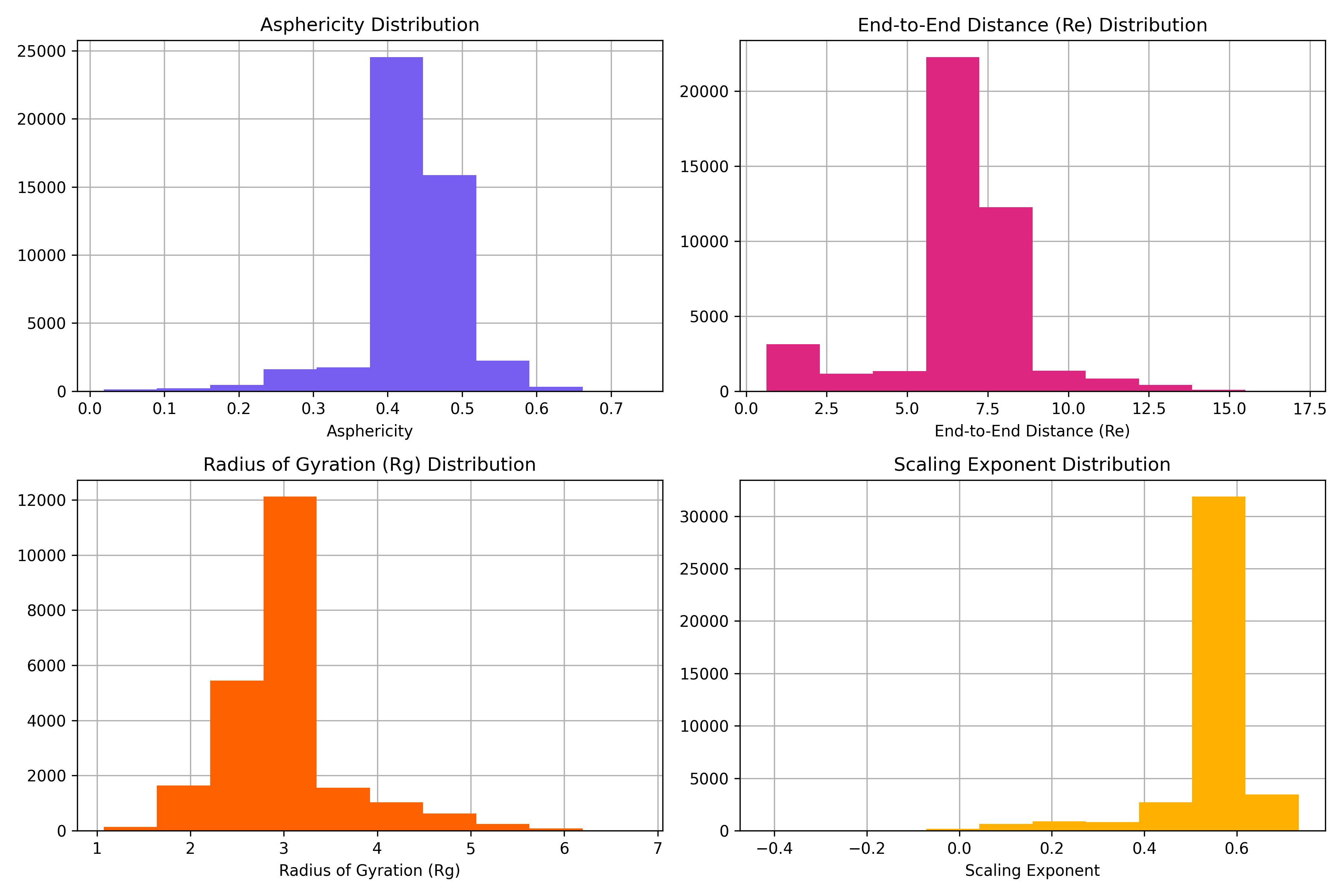
|
fuson_plm/benchmarking/idr_prediction/raw_data/asph_bio_synth_training_data_cleaned_05_09_2023.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/asph_nat_meth_test.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/scaled_re_bio_synth_training_data_cleaned_05_09_2023.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/scaled_re_nat_meth_test.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/scaled_rg_bio_synth_training_data_cleaned_05_09_2023.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/scaled_rg_nat_meth_test.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/scaling_exp_bio_synth_training_data_cleaned_05_09_2023.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/raw_data/scaling_exp_nat_meth_test.tsv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
fuson_plm/benchmarking/idr_prediction/results/final/asph_best_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:7eb01f34593fd5ae38d48ac392295ce81f5cd662671290bbbf854f9ed2d7db49
|
| 3 |
+
size 334
|
fuson_plm/benchmarking/idr_prediction/results/final/asph_hyperparam_screen_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:42aaa45520244fb734bc58ea7e7176d54d0087e28fbcbcbed09f796868207b28
|
| 3 |
+
size 2770
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/esm2_t33_650M_UR50D_asph_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/esm2_t33_650M_UR50D_asph_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4510bc0b24e5a81ef38f7baabe2ec31741d4bace254be9b949e1d4753edb5f4b
|
| 3 |
+
size 155775
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/fuson_plm_asph_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/asph/fuson_plm_asph_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:36357c139aff2d7a1ef869c4fa82b7f9a6bcf87298db7df59a8198a2c5b2d793
|
| 3 |
+
size 155818
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/esm2_t33_650M_UR50D_scaled_re_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/esm2_t33_650M_UR50D_scaled_re_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:64e6abb6c5b774ee9971885605c75bf6d0fec4aeea405b20c129ef0ecc21f1ef
|
| 3 |
+
size 176485
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/fuson_plm_scaled_re_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_re/fuson_plm_scaled_re_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:865b269369ce7903825d8225cb887ce226e2cd3b882a18c3bdad642cef769bd3
|
| 3 |
+
size 176364
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/esm2_t33_650M_UR50D_scaled_rg_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/esm2_t33_650M_UR50D_scaled_rg_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f362950ad91f14acffe261810bf6d7b2f484a16cff1ee31d1f2ea65f271a0725
|
| 3 |
+
size 93933
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/fuson_plm_scaled_rg_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaled_rg/fuson_plm_scaled_rg_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:469d1127b315f0e6b3091a644dff3d5bf658dae6daa0155cad70bd108132ba60
|
| 3 |
+
size 93867
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/esm2_t33_650M_UR50D_scaling_exp_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/esm2_t33_650M_UR50D_scaling_exp_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:44333a3310c7281e5e0b83bc90b2daec76ce13528c012d54f6773f4ee5fca91b
|
| 3 |
+
size 131908
|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/fuson_plm_scaling_exp_R2.png
ADDED

|
fuson_plm/benchmarking/idr_prediction/results/final/r2_plots/scaling_exp/fuson_plm_scaling_exp_R2_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:964cbefc62e9f4671e0502dd86fda8f079d78a2a5c637fa920bc023151169bf6
|
| 3 |
+
size 131764
|
fuson_plm/benchmarking/idr_prediction/results/final/scaled_re_best_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:82cfbb8a8bcf31e4bdf21e3fbe0454366677878f08fabcf141ac6b284936fe1d
|
| 3 |
+
size 343
|
fuson_plm/benchmarking/idr_prediction/results/final/scaled_re_hyperparam_screen_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:54b1f8503d37796b9463227cbe70b7b3a0a27f26274e409e3a2e04fa3c2f850a
|
| 3 |
+
size 2871
|
fuson_plm/benchmarking/idr_prediction/results/final/scaled_rg_best_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:460abf92fcad200cae609c973e96b1d4e56fe64cdb53449a9a3a22d64293716c
|
| 3 |
+
size 343
|
fuson_plm/benchmarking/idr_prediction/results/final/scaled_rg_hyperparam_screen_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8a728bf8f1f9b36a7d7b3ae49d24ad9eeb084aa342c01b87d72427442d730683
|
| 3 |
+
size 2867
|
fuson_plm/benchmarking/idr_prediction/results/final/scaling_exp_best_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:9de21dbfd32a9e5b751a61025b22a248485cf25d9e359174262e7984ecb35c17
|
| 3 |
+
size 346
|
fuson_plm/benchmarking/idr_prediction/results/final/scaling_exp_hyperparam_screen_test_r2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:763b89d2e1fb43fe855e42ed12da5786ebcd297fe5e31b2a238f852a22700eb6
|
| 3 |
+
size 2912
|
fuson_plm/benchmarking/idr_prediction/split.py
ADDED
|
@@ -0,0 +1,135 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from fuson_plm.utils.logging import open_logfile, log_update
|
| 2 |
+
from fuson_plm.utils.visualizing import set_font
|
| 3 |
+
from fuson_plm.benchmarking.idr_prediction.config import SPLIT
|
| 4 |
+
from fuson_plm.utils.splitting import split_clusters, check_split_validity
|
| 5 |
+
import os
|
| 6 |
+
import pandas as pd
|
| 7 |
+
|
| 8 |
+
def get_training_dfs(train, val, test, idr_db):
|
| 9 |
+
"""
|
| 10 |
+
Remove unnecessary columns for efficient storing of train, validation, and test sets for benchmarking.
|
| 11 |
+
Also, add the values using idr_db
|
| 12 |
+
"""
|
| 13 |
+
log_update('\nMaking dataframes for IDR prediction benchmark...')
|
| 14 |
+
|
| 15 |
+
# Delete cluster-related columns we don't need
|
| 16 |
+
train = train.drop(columns=['representative seq_id','member seq_id', 'member length', 'representative seq']).rename(columns={'member seq':'Sequence'})
|
| 17 |
+
val = val.drop(columns=['representative seq_id','member seq_id', 'member length', 'representative seq']).rename(columns={'member seq':'Sequence'})
|
| 18 |
+
test = test.drop(columns=['representative seq_id','member seq_id', 'member length', 'representative seq']).rename(columns={'member seq':'Sequence'})
|
| 19 |
+
|
| 20 |
+
# Add values and make one df for each one
|
| 21 |
+
# idr_db values are in columns: asph,scaled_re,scaled_rg,scaling_exp
|
| 22 |
+
value_cols = ['asph','scaled_re','scaled_rg','scaling_exp']
|
| 23 |
+
return_dict = {}
|
| 24 |
+
for col in value_cols:
|
| 25 |
+
temp_train = pd.merge(train, idr_db[['Sequence',col]], on='Sequence',how='left').rename(columns={col:'Value'}).dropna(subset='Value')
|
| 26 |
+
temp_val = pd.merge(val, idr_db[['Sequence',col]], on='Sequence',how='left').rename(columns={col:'Value'}).dropna(subset='Value')
|
| 27 |
+
temp_test = pd.merge(test, idr_db[['Sequence',col]], on='Sequence',how='left').rename(columns={col:'Value'}).dropna(subset='Value')
|
| 28 |
+
return_dict[col] = {
|
| 29 |
+
'train': temp_train,
|
| 30 |
+
'val': temp_val,
|
| 31 |
+
'test': temp_test
|
| 32 |
+
}
|
| 33 |
+
|
| 34 |
+
return return_dict
|
| 35 |
+
|
| 36 |
+
def main():
|
| 37 |
+
"""
|
| 38 |
+
"""
|
| 39 |
+
# Read all the input files
|
| 40 |
+
LOG_PATH = "splitting_log.txt"
|
| 41 |
+
IDR_DB_PATH = SPLIT.IDR_DB_PATH
|
| 42 |
+
CLUSTER_OUTPUT_PATH = SPLIT.CLUSTER_OUTPUT_PATH
|
| 43 |
+
RANDOM_STATE_1 = SPLIT.RANDOM_STATE_1
|
| 44 |
+
TEST_SIZE_1 = SPLIT.TEST_SIZE_1
|
| 45 |
+
RANDOM_STATE_2 = SPLIT.RANDOM_STATE_2
|
| 46 |
+
TEST_SIZE_2 = SPLIT.TEST_SIZE_2
|
| 47 |
+
|
| 48 |
+
# set font
|
| 49 |
+
set_font()
|
| 50 |
+
|
| 51 |
+
# Prepare the log file
|
| 52 |
+
with open_logfile(LOG_PATH):
|
| 53 |
+
log_update("Loaded data-splitting configurations from config.py")
|
| 54 |
+
SPLIT.print_config(indent='\t')
|
| 55 |
+
|
| 56 |
+
# Prepare directory to save results
|
| 57 |
+
os.makedirs("splits",exist_ok=True)
|
| 58 |
+
|
| 59 |
+
# Read the clusters and get a list of the representative IDs for splitting
|
| 60 |
+
clusters = pd.read_csv(CLUSTER_OUTPUT_PATH)
|
| 61 |
+
reps = clusters['representative seq_id'].unique().tolist()
|
| 62 |
+
log_update(f"\nPreparing clusters...\n\tCollected {len(reps)} clusters for splitting")
|
| 63 |
+
|
| 64 |
+
# Make the splits and extract the results
|
| 65 |
+
splits = split_clusters(reps,
|
| 66 |
+
random_state_1 = RANDOM_STATE_1, test_size_1 = TEST_SIZE_1,
|
| 67 |
+
random_state_2= RANDOM_STATE_2, test_size_2 = TEST_SIZE_2)
|
| 68 |
+
X_train = splits['X_train']
|
| 69 |
+
X_val = splits['X_val']
|
| 70 |
+
X_test = splits['X_test']
|
| 71 |
+
|
| 72 |
+
# Make slices of clusters dataframe for train, val, and test
|
| 73 |
+
train_clusters = clusters.loc[clusters['representative seq_id'].isin(X_train)].reset_index(drop=True)
|
| 74 |
+
val_clusters = clusters.loc[clusters['representative seq_id'].isin(X_val)].reset_index(drop=True)
|
| 75 |
+
test_clusters = clusters.loc[clusters['representative seq_id'].isin(X_test)].reset_index(drop=True)
|
| 76 |
+
|
| 77 |
+
# Check validity
|
| 78 |
+
check_split_validity(train_clusters, val_clusters, test_clusters)
|
| 79 |
+
|
| 80 |
+
# Print min and max sequence lengths
|
| 81 |
+
min_train_seqlen = min(train_clusters['member seq'].str.len())
|
| 82 |
+
max_train_seqlen = max(train_clusters['member seq'].str.len())
|
| 83 |
+
min_val_seqlen = min(val_clusters['member seq'].str.len())
|
| 84 |
+
max_val_seqlen = max(val_clusters['member seq'].str.len())
|
| 85 |
+
min_test_seqlen = min(test_clusters['member seq'].str.len())
|
| 86 |
+
max_test_seqlen = max(test_clusters['member seq'].str.len())
|
| 87 |
+
log_update(f"\nLength breakdown summary...\n\tTrain: min seq length = {min_train_seqlen}, max seq length = {max_train_seqlen}")
|
| 88 |
+
log_update(f"\nVal: min seq length = {min_val_seqlen}, max seq length = {max_val_seqlen}")
|
| 89 |
+
log_update(f"\nTest: min seq length = {min_test_seqlen}, max seq length = {max_test_seqlen}")
|
| 90 |
+
|
| 91 |
+
# cols = representative seq_id,member seq_id,representative seq,member seq
|
| 92 |
+
train_clusters.to_csv("splits/train_cluster_split.csv",index=False)
|
| 93 |
+
val_clusters.to_csv("splits/val_cluster_split.csv",index=False)
|
| 94 |
+
test_clusters.to_csv("splits/test_cluster_split.csv",index=False)
|
| 95 |
+
log_update('\nSaved cluster splits to splits/train_cluster_split.csv, splits/val_cluster_split.csv, splits/test_cluster_split.csv')
|
| 96 |
+
cols=','.join(list(train_clusters.columns))
|
| 97 |
+
log_update(f'\tColumns: {cols}')
|
| 98 |
+
|
| 99 |
+
# Get final dataframes for training, and check their distributions
|
| 100 |
+
idr_db = pd.read_csv(IDR_DB_PATH)
|
| 101 |
+
train_dfs_dict = get_training_dfs(train_clusters, val_clusters, test_clusters, idr_db)
|
| 102 |
+
os.makedirs('splits',exist_ok=True)
|
| 103 |
+
train_test_values_dict = {}
|
| 104 |
+
idr_property_name_dict = {'asph':'Asphericity','scaled_re':'End-to-End Distance (Re)','scaled_rg':'Radius of Gyration (Rg)','scaling_exp':'Scaling Exponent'}
|
| 105 |
+
|
| 106 |
+
for idr_property, dfs in train_dfs_dict.items():
|
| 107 |
+
os.makedirs(f"splits/{idr_property}", exist_ok=True)
|
| 108 |
+
train_df = dfs['train']
|
| 109 |
+
val_df = dfs['val']
|
| 110 |
+
test_df = dfs['test']
|
| 111 |
+
|
| 112 |
+
total_seqs = len(train_df)+len(val_df)+len(test_df)
|
| 113 |
+
train_df.to_csv(f"splits/{idr_property}/train_df.csv",index=False)
|
| 114 |
+
val_df.to_csv(f"splits/{idr_property}/val_df.csv",index=False)
|
| 115 |
+
test_df.to_csv(f"splits/{idr_property}/test_df.csv",index=False)
|
| 116 |
+
log_update(f"\nSaved {idr_property} training dataframes to splits/{idr_property}/train_df.csv, splits/{idr_property}/val_df.csv splits/test_df.csv")
|
| 117 |
+
log_update(f"\tTrain sequences: {len(train_df)} ({100*len(train_df)/total_seqs:.2f}%)")
|
| 118 |
+
log_update(f"\tVal sequences: {len(val_df)} ({100*len(val_df)/total_seqs:.2f}%)")
|
| 119 |
+
log_update(f"\tTest sequences: {len(test_df)} ({100*len(test_df)/total_seqs:.2f}%)")
|
| 120 |
+
log_update(f"\tTotal: {total_seqs}")
|
| 121 |
+
|
| 122 |
+
# Make sure the lengths are right
|
| 123 |
+
log_update(len(idr_db[idr_db[idr_property].notna()]))
|
| 124 |
+
assert total_seqs == len(idr_db[idr_db[idr_property].notna()])
|
| 125 |
+
|
| 126 |
+
train_test_values_dict[
|
| 127 |
+
idr_property_name_dict[idr_property]
|
| 128 |
+
] = {
|
| 129 |
+
'train': train_df['Value'].tolist(),
|
| 130 |
+
'val': val_df['Value'].tolist(),
|
| 131 |
+
'test': test_df['Value'].tolist()
|
| 132 |
+
}
|
| 133 |
+
|
| 134 |
+
if __name__ == "__main__":
|
| 135 |
+
main()
|
fuson_plm/benchmarking/idr_prediction/splits/asph/test_df.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:fa7f71e8371fab3cea404394674f551a720b0880fd977811b9729f46c09ab3eb
|
| 3 |
+
size 660037
|
fuson_plm/benchmarking/idr_prediction/splits/asph/train_df.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:bf500418ec8fea09477a25e44c385d1e0163b4089b21e63ab3002dbe3e0679a8
|
| 3 |
+
size 5235827
|
fuson_plm/benchmarking/idr_prediction/splits/asph/val_df.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:011ca834d11d0a2bf024af0e241032562d67fe9241e5e337e6a1739a468611fa
|
| 3 |
+
size 653940
|
fuson_plm/benchmarking/idr_prediction/splits/scaled_re/test_df.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:840c2174c82f83d2f6f2e994d94eef569379f79c1e1a837563a79489e65a2685
|
| 3 |
+
size 582519
|