Yuning You
commited on
Commit
·
eaae86c
1
Parent(s):
e100fb6
update
Browse files- README.md +3 -2
- figures/autoregressive.gif +0 -0
README.md
CHANGED
|
@@ -15,8 +15,8 @@ tags:
|
|
| 15 |
## Overview
|
| 16 |
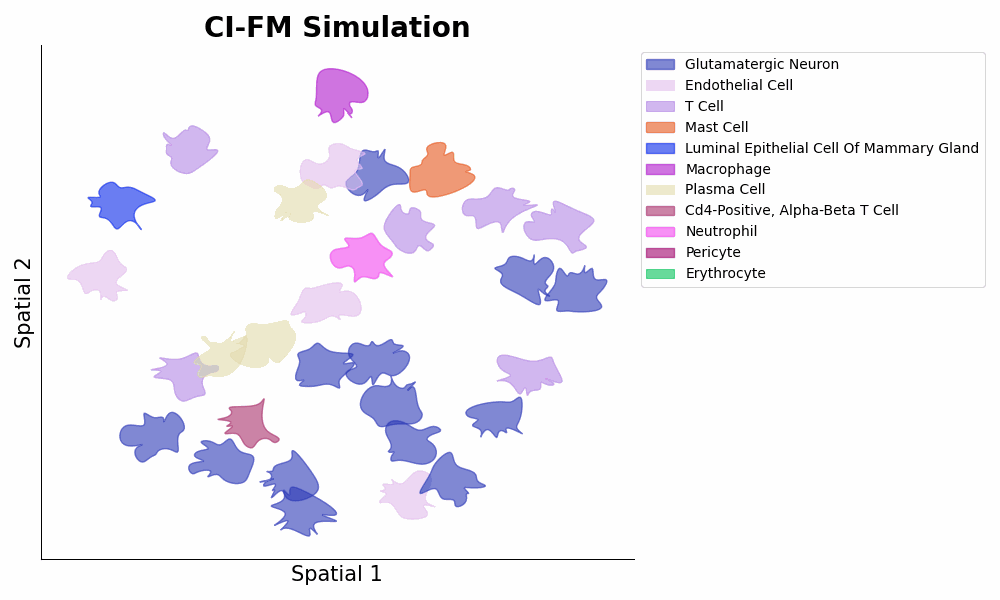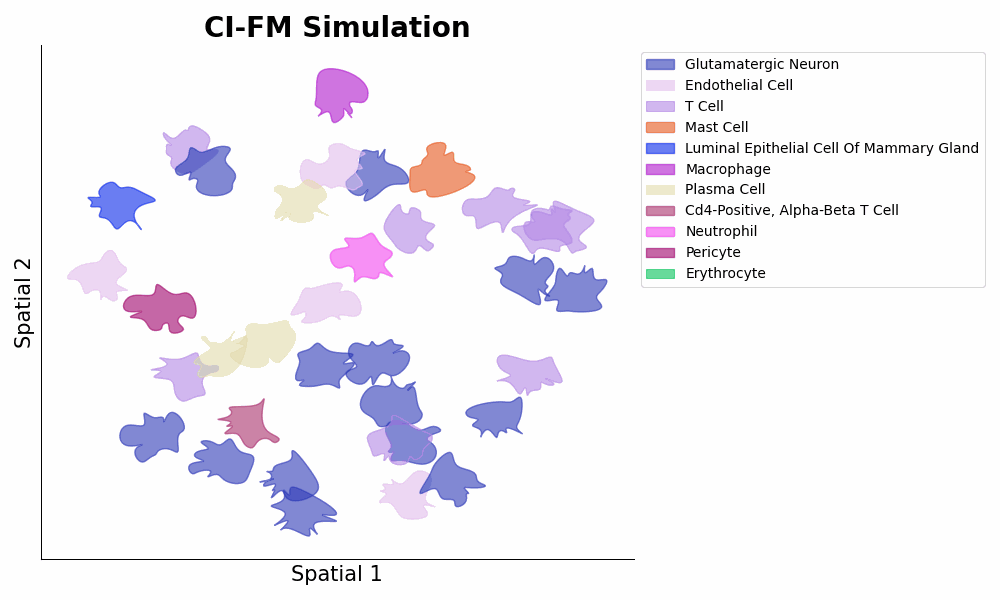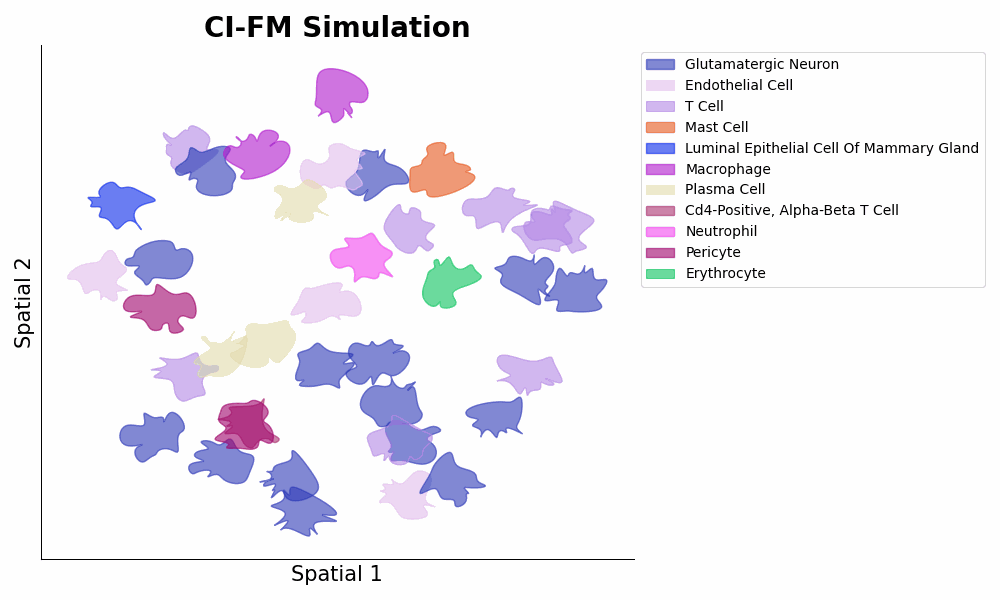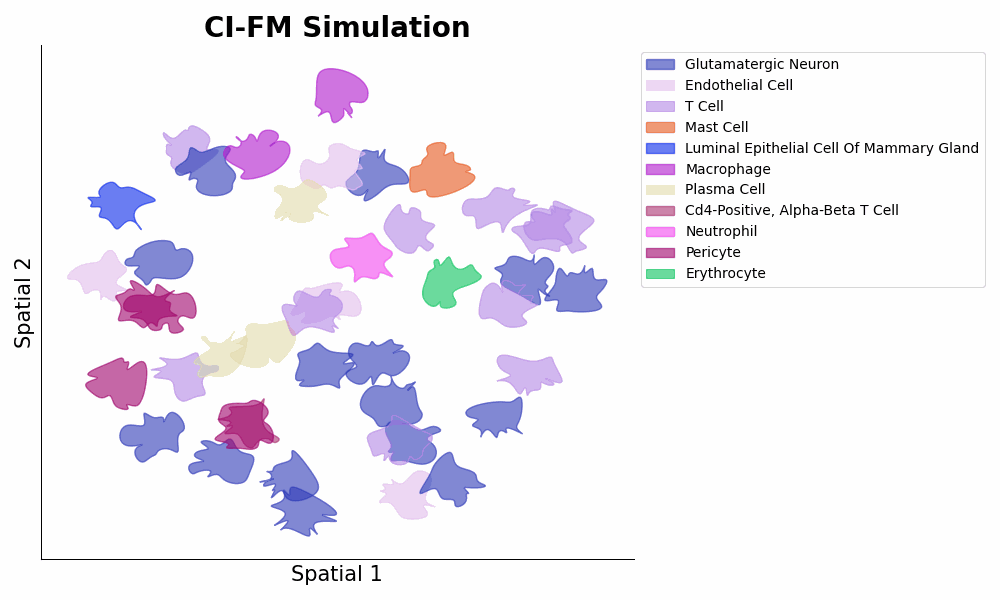
This is the PyTorch implementation of the CI-FM model -- an AI model that can simulate the activities within a living tissue (AI virtual tissue).
|
| 17 |
The current version of CI-FM has 138M parameters and is trained on around 23M cells of spatial genomics. The signature functions of CI-FM are:
|
| 18 |
-
- **Embedding** of celllular microenvironments via ```embeddings = model.embed(adata)``` (Figure below panel D top);
|
| 19 |
-
- **Inference** of cellular gene expressions within a certain microenvironment via ```expressions = model.predict_cells_at_locations(adata, target_locs)``` (Figure below panel D bottom).
|
| 20 |
|
| 21 |
The detailed usage of the model can be found in the [tutorial](https://huggingface.co/ynyou/CIFM/blob/main/test.ipynb).
|
| 22 |
Before running the tutorial, please set up an environment following the [environment instruction](https://huggingface.co/ynyou/CIFM#environment).
|
|
@@ -24,6 +24,7 @@ Before running the tutorial, please set up an environment following the [environ
|
|
| 24 |
More information about the model can be found in the [preprint]().
|
| 25 |
|
| 26 |

|
|
|
|
| 27 |
|
| 28 |
## Environment
|
| 29 |
I use conda to manage the environment ```conda create -n $MYENV python=3.11```, but it is not the only way to do that.
|
|
|
|
| 15 |
## Overview
|
| 16 |
This is the PyTorch implementation of the CI-FM model -- an AI model that can simulate the activities within a living tissue (AI virtual tissue).
|
| 17 |
The current version of CI-FM has 138M parameters and is trained on around 23M cells of spatial genomics. The signature functions of CI-FM are:
|
| 18 |
+
- **Embedding** of celllular microenvironments via ```embeddings = model.embed(adata)``` (1st Figure below panel D top);
|
| 19 |
+
- **Inference** of cellular gene expressions within a certain microenvironment via ```expressions = model.predict_cells_at_locations(adata, target_locs)``` (1st Figure below panel D bottom, and 2nd Figure below).
|
| 20 |
|
| 21 |
The detailed usage of the model can be found in the [tutorial](https://huggingface.co/ynyou/CIFM/blob/main/test.ipynb).
|
| 22 |
Before running the tutorial, please set up an environment following the [environment instruction](https://huggingface.co/ynyou/CIFM#environment).
|
|
|
|
| 24 |
More information about the model can be found in the [preprint]().
|
| 25 |
|
| 26 |

|
| 27 |
+

|
| 28 |
|
| 29 |
## Environment
|
| 30 |
I use conda to manage the environment ```conda create -n $MYENV python=3.11```, but it is not the only way to do that.
|
figures/autoregressive.gif
ADDED
|