Spaces:
Sleeping
Sleeping
Reorganized folders.
Browse filesAdded training graph.
app.py
CHANGED
|
@@ -14,12 +14,12 @@ examples_path = "examples"
|
|
| 14 |
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
|
| 15 |
|
| 16 |
correct_preds, wrong_preds = {}, {}
|
| 17 |
-
condition_lst = pd.read_csv("feature.csv", header = "infer", sep = ",", encoding = "utf-8", dtype=str)
|
| 18 |
-
D_LABITEMS = pd.read_csv("D_LABITEMS.csv", header = "infer", sep = ",", encoding = "utf-8", dtype=str)
|
| 19 |
|
| 20 |
|
| 21 |
def load_model():
|
| 22 |
-
path = r"final_model.pt"
|
| 23 |
kwargs, state = torch.load(path, weights_only=False, map_location=device)
|
| 24 |
model = VariationalGNN(**kwargs).to(device)
|
| 25 |
model.load_state_dict(state)
|
|
@@ -236,7 +236,7 @@ with gr.Blocks(theme=gr.themes.Soft(), css=css) as demo:
|
|
| 236 |
query_tbx.change(fn=query_input_change_event, inputs=[query_tbx, query_type], outputs=[query_btn, html])
|
| 237 |
query_type.change(fn=query_input_change_event, inputs=[query_tbx, query_type], outputs=[query_btn, html])
|
| 238 |
query_btn.click(fn=do_query, inputs=[query_tbx, query_type], outputs=html)
|
| 239 |
-
with gr.Accordion("More on
|
| 240 |
gr.Markdown(
|
| 241 |
"""
|
| 242 |
- Paper: [Variationally Regularized Graph-based Representation Learning for Electronic Health Records (Zhu et al, 2021)](https://arxiv.org/abs/1912.03761)
|
|
@@ -250,9 +250,13 @@ with gr.Blocks(theme=gr.themes.Soft(), css=css) as demo:
|
|
| 250 |
- Trained on NVIDIA A100 with PyTorch 2.4.0
|
| 251 |
"""
|
| 252 |
)
|
|
|
|
|
|
|
|
|
|
|
|
|
| 253 |
|
| 254 |
for partialFunc in resDispPartFuncs:
|
| 255 |
partialFunc(outputs=[result_pred, result_label])
|
| 256 |
|
| 257 |
|
| 258 |
-
demo.launch(debug=True)
|
|
|
|
| 14 |
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
|
| 15 |
|
| 16 |
correct_preds, wrong_preds = {}, {}
|
| 17 |
+
condition_lst = pd.read_csv("data/feature.csv", header = "infer", sep = ",", encoding = "utf-8", dtype=str)
|
| 18 |
+
D_LABITEMS = pd.read_csv("data/D_LABITEMS.csv", header = "infer", sep = ",", encoding = "utf-8", dtype=str)
|
| 19 |
|
| 20 |
|
| 21 |
def load_model():
|
| 22 |
+
path = r"models/final_model.pt"
|
| 23 |
kwargs, state = torch.load(path, weights_only=False, map_location=device)
|
| 24 |
model = VariationalGNN(**kwargs).to(device)
|
| 25 |
model.load_state_dict(state)
|
|
|
|
| 236 |
query_tbx.change(fn=query_input_change_event, inputs=[query_tbx, query_type], outputs=[query_btn, html])
|
| 237 |
query_type.change(fn=query_input_change_event, inputs=[query_tbx, query_type], outputs=[query_btn, html])
|
| 238 |
query_btn.click(fn=do_query, inputs=[query_tbx, query_type], outputs=html)
|
| 239 |
+
with gr.Accordion("More on Technical Details...", open=False):
|
| 240 |
gr.Markdown(
|
| 241 |
"""
|
| 242 |
- Paper: [Variationally Regularized Graph-based Representation Learning for Electronic Health Records (Zhu et al, 2021)](https://arxiv.org/abs/1912.03761)
|
|
|
|
| 250 |
- Trained on NVIDIA A100 with PyTorch 2.4.0
|
| 251 |
"""
|
| 252 |
)
|
| 253 |
+
with gr.Accordion("More on Training...", open=False):
|
| 254 |
+
gr.HTML("""
|
| 255 |
+
<img src="/file=images/AUPRC_Training_Graph.png" alt="">
|
| 256 |
+
""")
|
| 257 |
|
| 258 |
for partialFunc in resDispPartFuncs:
|
| 259 |
partialFunc(outputs=[result_pred, result_label])
|
| 260 |
|
| 261 |
|
| 262 |
+
demo.launch(debug=True, allowed_paths=["images/."])
|
D_LABITEMS.csv → data/D_LABITEMS.csv
RENAMED
|
File without changes
|
feature.csv → data/feature.csv
RENAMED
|
File without changes
|
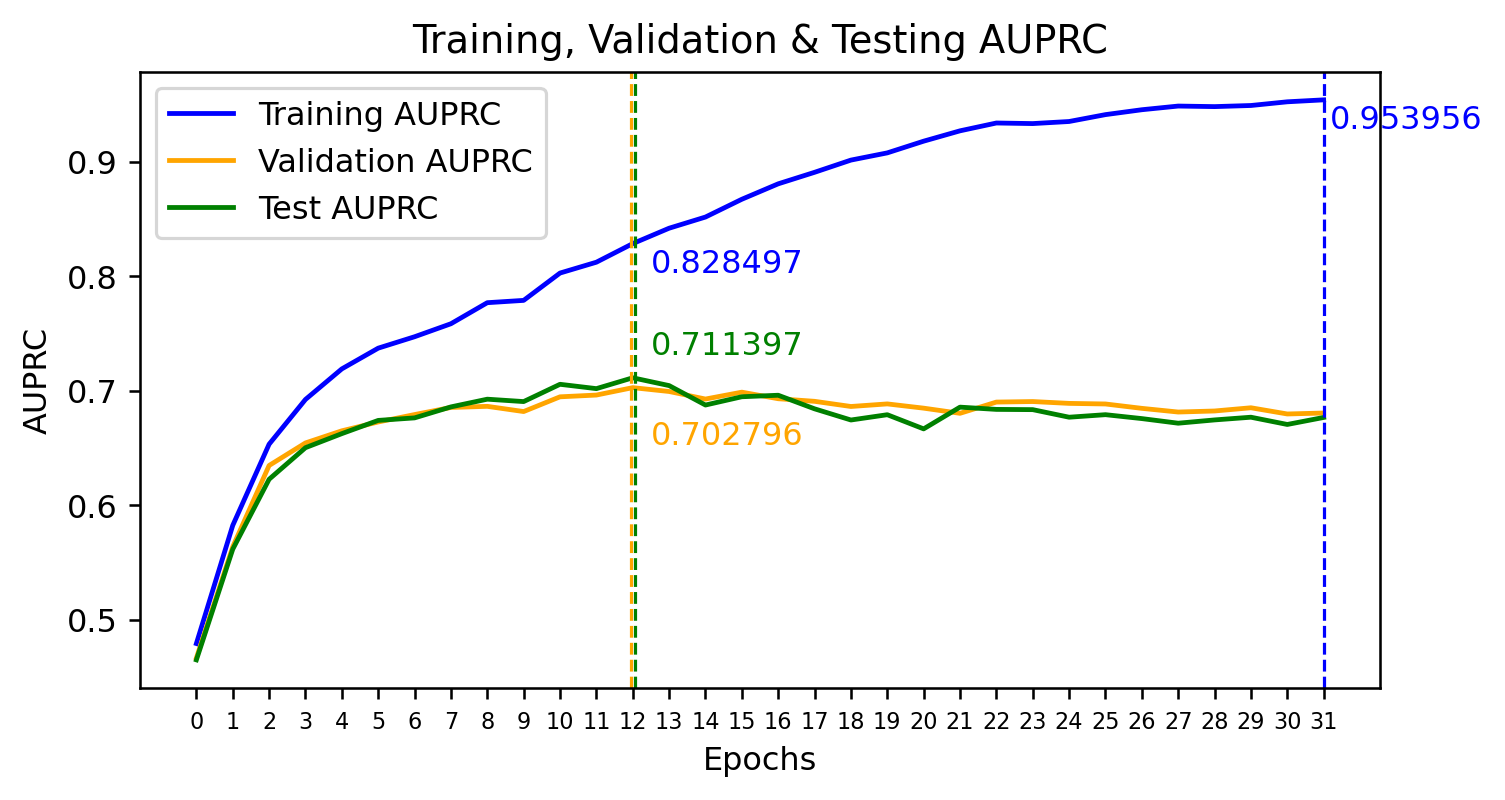
images/AUPRC_Training_Graph.png
ADDED
|
final_model.pt → models/final_model.pt
RENAMED
|
File without changes
|