Spaces:
Sleeping
Sleeping
Trương Gia Bảo
commited on
Commit
·
30872a6
1
Parent(s):
4bc10ce
Update UI
Browse files- .gitattributes +1 -0
- about.png +0 -0
- app.py +50 -12
- img/about1.png +3 -0
- img/about2.png +3 -0
- img/about3.png +3 -0
- img/model_fig.png +3 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 35 |
model.iter-685000 filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 35 |
model.iter-685000 filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
*.png filter=lfs diff=lfs merge=lfs -text
|
about.png
CHANGED
|
|
Git LFS Details
|
app.py
CHANGED
|
@@ -56,9 +56,9 @@ def img_to_bytes(img_path):
|
|
| 56 |
img_bytes = Path(img_path).read_bytes()
|
| 57 |
encoded = base64.b64encode(img_bytes).decode()
|
| 58 |
return encoded
|
| 59 |
-
def img_to_html(img_path):
|
| 60 |
-
img_html = "<img src='data:image/png;base64,{}' class='img-fluid' style='max-width:
|
| 61 |
-
img_to_bytes(img_path)
|
| 62 |
)
|
| 63 |
return img_html
|
| 64 |
|
|
@@ -276,8 +276,11 @@ def colorize_step(n_step,cur_step):
|
|
| 276 |
return color_list
|
| 277 |
|
| 278 |
def form_header():
|
| 279 |
-
st.markdown("<h1 style='text-align: center;'>Junction Tree Variational Autoencoder
|
| 280 |
-
st.markdown("<
|
|
|
|
|
|
|
|
|
|
| 281 |
|
| 282 |
# determines button color which should be red when user is on that given step
|
| 283 |
oam_type = 'primary' if st.session_state['current_view'] == 0 else 'secondary'
|
|
@@ -309,14 +312,49 @@ def form_body():
|
|
| 309 |
About()
|
| 310 |
|
| 311 |
def About():
|
| 312 |
-
|
| 313 |
We seek to automate the design of molecules based on specific chemical properties. In computational terms, this task involves continuous embedding and generation of molecular graphs. Our primary contribution is the direct realization of molecular graphs, a task previously approached by generating linear SMILES strings instead of graphs. Our junction tree variational autoencoder generates molecular graphs in two phases, by first generating a tree-structured scaffold over chemical substructures, and then combining them into a molecule with a graph message passing network. This approach allows us to incrementally expand molecules while maintaining chemical validity at every step. We evaluate our model on multiple tasks ranging from molecular generation to optimization. Across these tasks, our model outperforms previous state-of-the-art baselines by a significant margin.
|
| 314 |
-
|
| 315 |
-
|
| 316 |
-
|
| 317 |
-
|
| 318 |
-
|
| 319 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 320 |
|
| 321 |
def Optimize_a_molecule():
|
| 322 |
st.markdown("<h2 style='text-align: center;'>Optimize a molecule</h2>",unsafe_allow_html=True)
|
|
|
|
| 56 |
img_bytes = Path(img_path).read_bytes()
|
| 57 |
encoded = base64.b64encode(img_bytes).decode()
|
| 58 |
return encoded
|
| 59 |
+
def img_to_html(img_path,max_width=500):
|
| 60 |
+
img_html = "<img src='data:image/png;base64,{}' class='img-fluid' style='max-width: {}px;'>".format(
|
| 61 |
+
img_to_bytes(img_path), max_width
|
| 62 |
)
|
| 63 |
return img_html
|
| 64 |
|
|
|
|
| 276 |
return color_list
|
| 277 |
|
| 278 |
def form_header():
|
| 279 |
+
st.markdown("<h1 style='padding: 25px;text-align: center;color: white;background-color: tomato;'>Molecular Optimization using Junction Tree Variational Autoencoder</h1>",unsafe_allow_html=True)
|
| 280 |
+
st.markdown("<h4 style='padding: 10px;text-align: center;color: white;background-color: mediumseagreen;'>Gia-Bao Truong</h4>",unsafe_allow_html=True)
|
| 281 |
+
with st.expander(':star2: About the model'):
|
| 282 |
+
st.markdown("<p style='text-align: center;'>Based on Junction Tree Variational Autoencoder for Molecular Graph Generation (JTVAE)</p>",unsafe_allow_html=True)
|
| 283 |
+
st.markdown("<p style='text-align: center;'>Wengong Jin, Regina Barzilay, Tommi Jaakkola</p>",unsafe_allow_html=True)
|
| 284 |
|
| 285 |
# determines button color which should be red when user is on that given step
|
| 286 |
oam_type = 'primary' if st.session_state['current_view'] == 0 else 'secondary'
|
|
|
|
| 312 |
About()
|
| 313 |
|
| 314 |
def About():
|
| 315 |
+
descrip_model = '''
|
| 316 |
We seek to automate the design of molecules based on specific chemical properties. In computational terms, this task involves continuous embedding and generation of molecular graphs. Our primary contribution is the direct realization of molecular graphs, a task previously approached by generating linear SMILES strings instead of graphs. Our junction tree variational autoencoder generates molecular graphs in two phases, by first generating a tree-structured scaffold over chemical substructures, and then combining them into a molecule with a graph message passing network. This approach allows us to incrementally expand molecules while maintaining chemical validity at every step. We evaluate our model on multiple tasks ranging from molecular generation to optimization. Across these tasks, our model outperforms previous state-of-the-art baselines by a significant margin.
|
| 317 |
+
'''
|
| 318 |
+
img_caption = '''
|
| 319 |
+
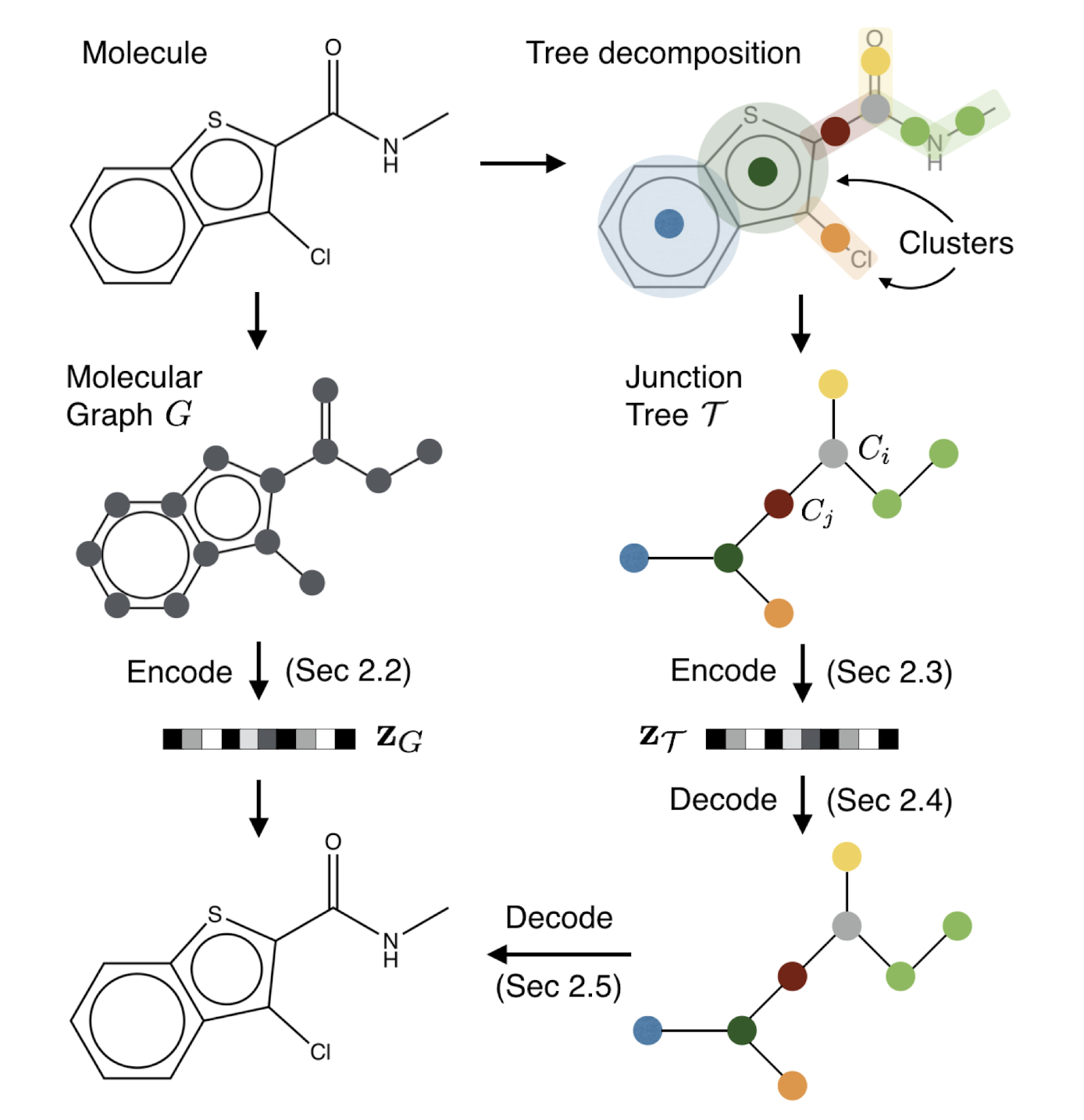
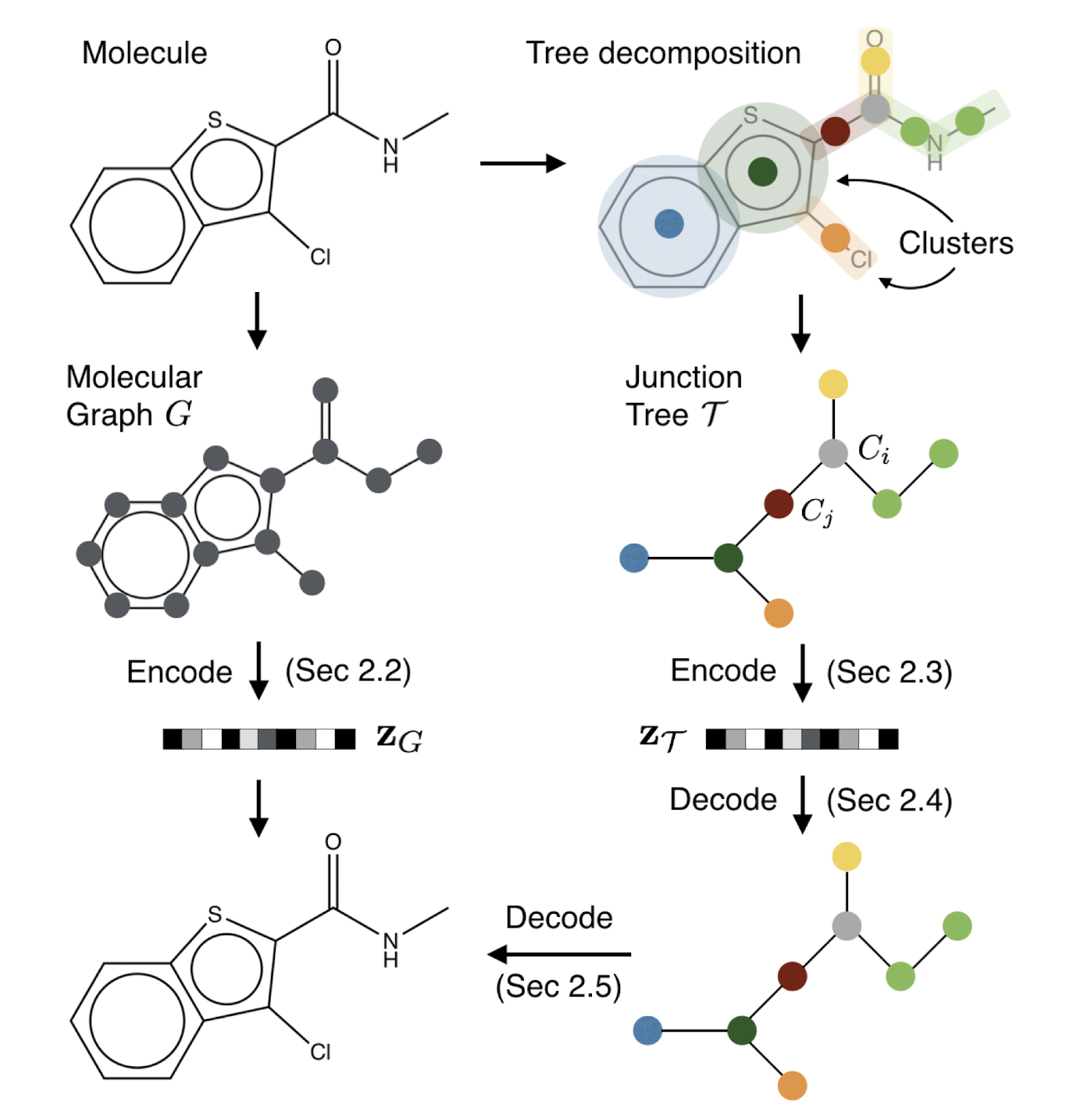
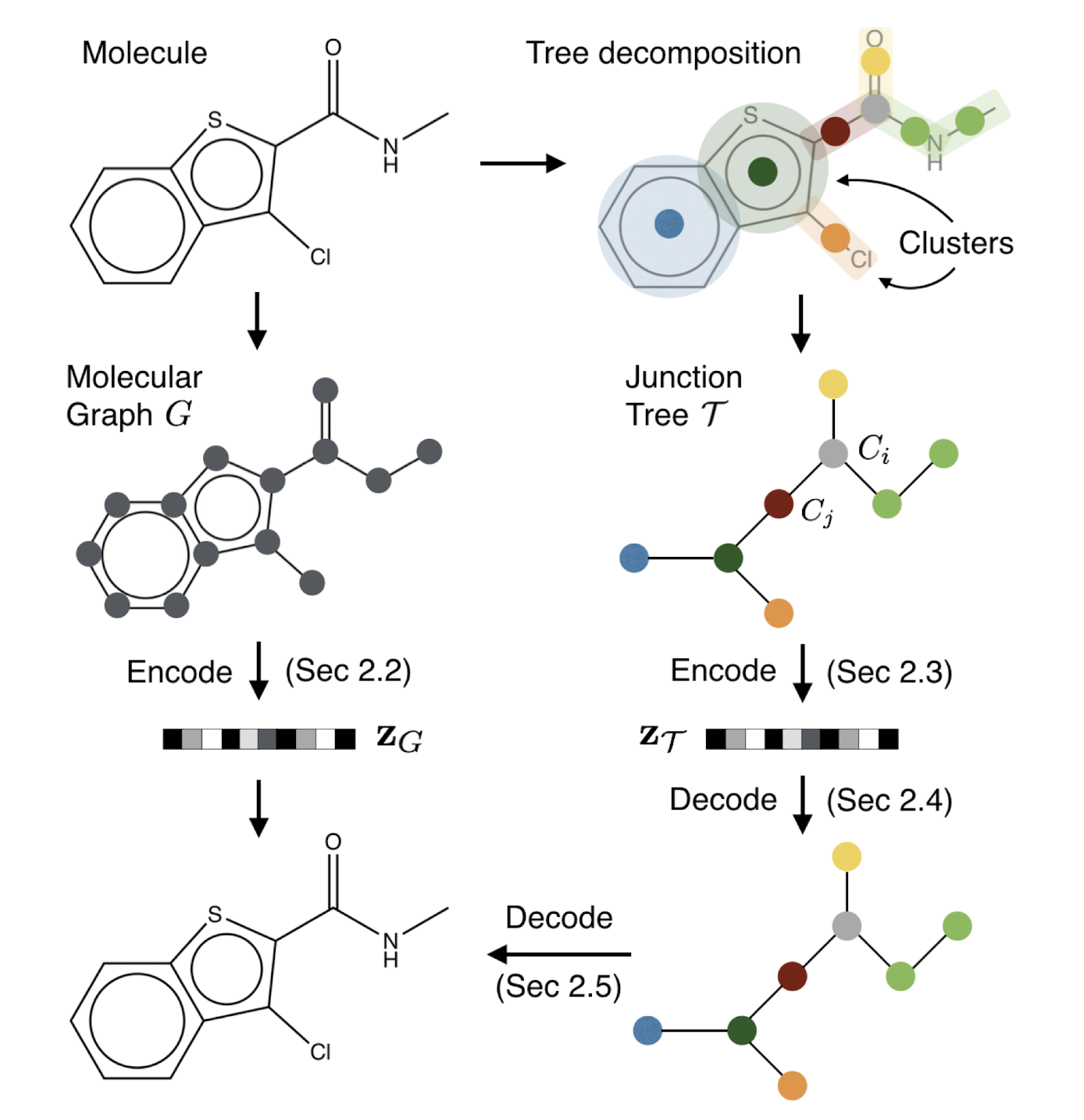
Figure 3. Overview of our method: A molecular graph G is first decomposed into its junction tree TG, where each colored node in the tree represents a substructure in the molecule. We then encode both the tree and graph into their latent embeddings zT and zG. To decode the molecule, we first reconstruct junction tree from zT , and then assemble nodes in the tree back to the original molecule.'''
|
| 320 |
+
|
| 321 |
+
with st.expander(':four_leaf_clover: About the author',expanded=True):
|
| 322 |
+
st.markdown("<h4 style='text-align:center;'>Gia-Bao Truong</h4>",unsafe_allow_html=True)
|
| 323 |
+
st.markdown("<h4 style='color:tomato; text-align:center;'>Student at</h4>",unsafe_allow_html=True)
|
| 324 |
+
st.markdown("<p style='text-align:center;'>"+
|
| 325 |
+
img_to_html('img/about1.png',64)+' '+img_to_html('img/about2.png',64)+
|
| 326 |
+
"</p>", unsafe_allow_html=True)
|
| 327 |
+
st.markdown("<h5 style='text-align:center;'>Faculty of Pharmacy, University of Medicine and Pharmacy at Ho Chi Minh City</h5>",unsafe_allow_html=True)
|
| 328 |
+
st.markdown("<h4 style='color:tomato; text-align:center;'>Team</h4>",unsafe_allow_html=True)
|
| 329 |
+
st.markdown("<p style='text-align:center;'>"+
|
| 330 |
+
img_to_html('img/about3.png',64)+
|
| 331 |
+
"</p>", unsafe_allow_html=True)
|
| 332 |
+
st.markdown("<h5 style='text-align:center;'>MedAI</h5>",unsafe_allow_html=True)
|
| 333 |
+
|
| 334 |
+
|
| 335 |
+
with st.expander(':star2: About the model',expanded=True):
|
| 336 |
+
st.markdown("Based on Junction Tree Variational Autoencoder for Molecular Graph Generation (JTVAE)",unsafe_allow_html=True)
|
| 337 |
+
st.markdown("<h4 style='color:tomato;'>Citing</h4>",unsafe_allow_html=True)
|
| 338 |
+
st.markdown("Paper: [https://arxiv.org/abs/1802.04364](https://arxiv.org/abs/1802.04364)")
|
| 339 |
+
st.code('''@misc{jin2019junction,
|
| 340 |
+
title={Junction Tree Variational Autoencoder for Molecular Graph Generation},
|
| 341 |
+
author={Wengong Jin and Regina Barzilay and Tommi Jaakkola},
|
| 342 |
+
year={2019},
|
| 343 |
+
eprint={1802.04364},
|
| 344 |
+
archivePrefix={arXiv},
|
| 345 |
+
primaryClass={cs.LG}
|
| 346 |
+
}''')
|
| 347 |
+
st.markdown("<h4 style='color:tomato;'>Author</h4>",unsafe_allow_html=True)
|
| 348 |
+
st.markdown("Wengong Jin, Regina Barzilay, Tommi Jaakkola",unsafe_allow_html=True)
|
| 349 |
+
st.markdown("<h4 style='color:tomato;'>Abstract</h4>",unsafe_allow_html=True)
|
| 350 |
+
st.markdown(descrip_model)
|
| 351 |
+
ab = st.columns([1,10,1])
|
| 352 |
+
ab[1].markdown("<p style='text-align: center;'>"+
|
| 353 |
+
img_to_html('img/model_fig.png')+
|
| 354 |
+
"</p>", unsafe_allow_html=True)
|
| 355 |
+
ab[1].markdown("<p style='text-align: center;'>"+
|
| 356 |
+
img_caption+
|
| 357 |
+
"</p>",unsafe_allow_html=True)
|
| 358 |
|
| 359 |
def Optimize_a_molecule():
|
| 360 |
st.markdown("<h2 style='text-align: center;'>Optimize a molecule</h2>",unsafe_allow_html=True)
|
img/about1.png
ADDED

|
Git LFS Details
|
img/about2.png
ADDED

|
Git LFS Details
|
img/about3.png
ADDED

|
Git LFS Details
|
img/model_fig.png
ADDED
|
Git LFS Details
|