puncta benchmark
Browse files- fuson_plm/benchmarking/puncta/FOdb_physicochemical_embeddings.pkl +3 -0
- fuson_plm/benchmarking/puncta/README.md +95 -0
- fuson_plm/benchmarking/puncta/__init__.py +0 -0
- fuson_plm/benchmarking/puncta/clean.py +184 -0
- fuson_plm/benchmarking/puncta/cleaned_dataset_s4.csv +3 -0
- fuson_plm/benchmarking/puncta/cleaning_log.txt +3 -0
- fuson_plm/benchmarking/puncta/config.py +17 -0
- fuson_plm/benchmarking/puncta/plot.py +244 -0
- fuson_plm/benchmarking/puncta/results/final/cytoplasm_verificationFOs_results.csv +3 -0
- fuson_plm/benchmarking/puncta/results/final/figures/cytoplasm_verificationFOs_barchart.png +0 -0
- fuson_plm/benchmarking/puncta/results/final/figures/cytoplasm_verificationFOs_barchart_source_data.csv +3 -0
- fuson_plm/benchmarking/puncta/results/final/figures/formation_verificationFOs_0.83thresh_barchart.png +0 -0
- fuson_plm/benchmarking/puncta/results/final/figures/formation_verificationFOs_0.83thresh_barchart_source_data.csv +3 -0
- fuson_plm/benchmarking/puncta/results/final/figures/nucleus_verificationFOs_barchart.png +0 -0
- fuson_plm/benchmarking/puncta/results/final/figures/nucleus_verificationFOs_barchart_source_data.csv +3 -0
- fuson_plm/benchmarking/puncta/results/final/formation_verificationFOs_0.83thresh_results.csv +3 -0
- fuson_plm/benchmarking/puncta/results/final/nucleus_verificationFOs_results.csv +3 -0
- fuson_plm/benchmarking/puncta/splits.csv +3 -0
- fuson_plm/benchmarking/puncta/train.py +155 -0
fuson_plm/benchmarking/puncta/FOdb_physicochemical_embeddings.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:3d78986f0724138ed83c72fd4274154ca3f96a09f5fd8ad94030493375788006
|
| 3 |
+
size 168405
|
fuson_plm/benchmarking/puncta/README.md
ADDED
|
@@ -0,0 +1,95 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
## Puncta Prediction Benchmark
|
| 2 |
+
|
| 3 |
+
This folder contains all the data and code needed to perform the **puncta prediction benchmark** (Figure 3).
|
| 4 |
+
|
| 5 |
+
### From raw data to train/test splits
|
| 6 |
+
To train the puncta predictors, we processed raw data from FOdb [(Tripathi et al. 2023)](https://doi.org/10.1038/s41467-023-41655-2) Supplementary dataset 4 (`fuson_plm/data/raw_data/FOdb_puncta.csv`) and Supplementary dataset 5 (`fuson_plm/data/raw_data/FODb_SD5.csv`) using the file `clean.py` in the `puncta` directory.
|
| 7 |
+
|
| 8 |
+
```
|
| 9 |
+
data/
|
| 10 |
+
└── raw_data/
|
| 11 |
+
├── FOdb_puncta.csv
|
| 12 |
+
├── FOdb_SD5.csv
|
| 13 |
+
|
| 14 |
+
benchmarking/
|
| 15 |
+
└── puncta/
|
| 16 |
+
├── clean.py
|
| 17 |
+
├── cleaned_dataset_s4.csv
|
| 18 |
+
├── splits.csv
|
| 19 |
+
├── FOdb_physicochemical_embeddings.pkl
|
| 20 |
+
```
|
| 21 |
+
|
| 22 |
+
The `clean.py` script generates the following files:
|
| 23 |
+
- **`cleaned_dataset_s4.csv`**: clean version of `FOdb_puncta.csv`, where fusion oncoproteins with puncta status "Other" or "Nucleolar" have been removed, and only the 25 low-MI features from `FOdb_SD5.csv' are retained.
|
| 24 |
+
- **`splits.csv`**: fusion oncoproteins from `cleaned_dataset_s4.csv`, labeled in the `split` column as either being part of the *train* set ("Expressed_Set" in FOdb) or *test* set ("Verification_Set" in FOdb). This dataset also features `nucleus`, `cytoplasm`, and `formation` columns of 1s and 0s. In `nucleus`, 1=forms a condensate in the nucleus, 0=does not; in `cytoplasm`, 1=forms a condensate in the cytoplasm, 0=does not; in `formation`, 1=forms a condensate at all, 0=does not.
|
| 25 |
+
- **`FOdb_physicochemical_embeddings.pkl`**: a dictionary where fusion proteins from `splits.csv` are they keys, and their feature vectors of 25 low-MI features from `cleaned_dataset_s4.csv` are the values.
|
| 26 |
+
|
| 27 |
+
### Training
|
| 28 |
+
|
| 29 |
+
`config.py` holds training configuations.
|
| 30 |
+
|
| 31 |
+
```
|
| 32 |
+
# Benchmarking configs
|
| 33 |
+
BENCHMARK_FUSONPLM = True # True if you want to benchmark a FusOn-pLM Model
|
| 34 |
+
|
| 35 |
+
# FUSONPLM_CKPTS. If you've traiend your own model, this is a dictionary: key = run name, values = epochs
|
| 36 |
+
# If you want to use the trained FusOn-pLM, instead FUSONPLM_CKPTS="FusOn-pLM"
|
| 37 |
+
FUSONPLM_CKPTS= {}
|
| 38 |
+
|
| 39 |
+
# Model comparison configs
|
| 40 |
+
BENCHMARK_ESM = True # True if you want to benchmark ESM-2-650M
|
| 41 |
+
BENCHMARK_PROTT5 = True # True if you want to benchmark ProtT5
|
| 42 |
+
BENCHMARK_FO_PUNCTA_ML = True # True if you want to benchmark FO-Puncta-ML from the FOdb paper
|
| 43 |
+
|
| 44 |
+
# Overwriting configs
|
| 45 |
+
PERMISSION_TO_OVERWRITE = False # if False, script will halt if it believes these embeddings have already been made.
|
| 46 |
+
|
| 47 |
+
# GPU configs
|
| 48 |
+
CUDA_VISIBLE_DEVICES="0" # GPUs to make visible for this process
|
| 49 |
+
```
|
| 50 |
+
<br>
|
| 51 |
+
|
| 52 |
+
`train.py` will train the XGBoost classifiers.
|
| 53 |
+
- All **results** are stored in `puncta/results/timestamp`, where `timestamp` is a unique string encoding the date and time when you started training.
|
| 54 |
+
- All **embeddings** made for training will be stored in a new folder called `puncta/embeddings/` with subfolders for each model. This allows you to use the same model multiple times without regenerating embeddings.
|
| 55 |
+
|
| 56 |
+
```
|
| 57 |
+
benchmarking/
|
| 58 |
+
└── puncta/
|
| 59 |
+
└── embeddings/
|
| 60 |
+
└── esm2_t33_650M_UR50D/...
|
| 61 |
+
└── fuson_plm/...
|
| 62 |
+
└── prot_t5_xl_half_uniref50_enc/...
|
| 63 |
+
└── results/
|
| 64 |
+
└── final/
|
| 65 |
+
└── figures/
|
| 66 |
+
├── cytoplasm_verificationFOs_barchart_source_data.csv
|
| 67 |
+
├── cytoplasm_verificationFOs_barchart.png
|
| 68 |
+
├── formation_verificationFOs_0.83thresh_barchart_source_data.csv
|
| 69 |
+
├── formation_verificationFOs_0.83thresh_barchart.png
|
| 70 |
+
├── nucleus_verificationFOs_barchart_source_data.csv
|
| 71 |
+
├── nucleus_verificationFOs_barchart.png
|
| 72 |
+
├── cytoplasm_verificationFOs_results.csv
|
| 73 |
+
├── formation_verificationFOs_0.83thresh_results.csv
|
| 74 |
+
├── nucleus_verificationFOs_results.csv
|
| 75 |
+
```
|
| 76 |
+
|
| 77 |
+
The following files are in `results/final/figures`:
|
| 78 |
+
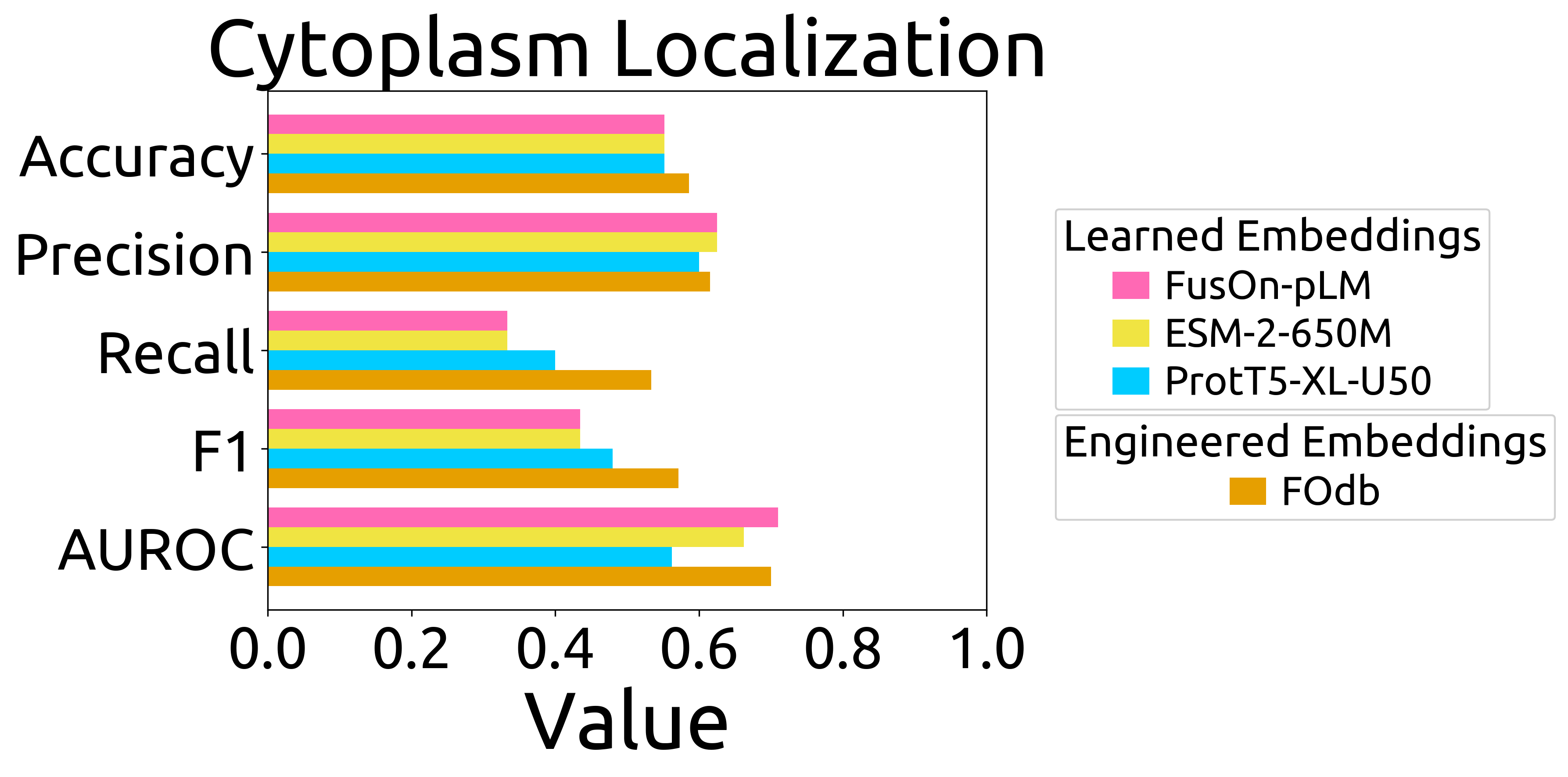
- **`cytoplasm_verificationFOs_barchart.png`**: bar chart of performance on the cytoplasm puncta prediction task (Fig. 3E), and the formatted data that went directly into the plot (`cytoplasm_verificationFOs_barchart_source_data.csv`)
|
| 79 |
+
- **`formation_verificationFOs_0.83thresh_barchart.png`**: bar chart of performance on the puncta formation prediction task (Fig. 3C), and the formatted data that went directly into the plot (`formation_verificationFOs_0.83thresh_barchart_source_data.csv`)
|
| 80 |
+
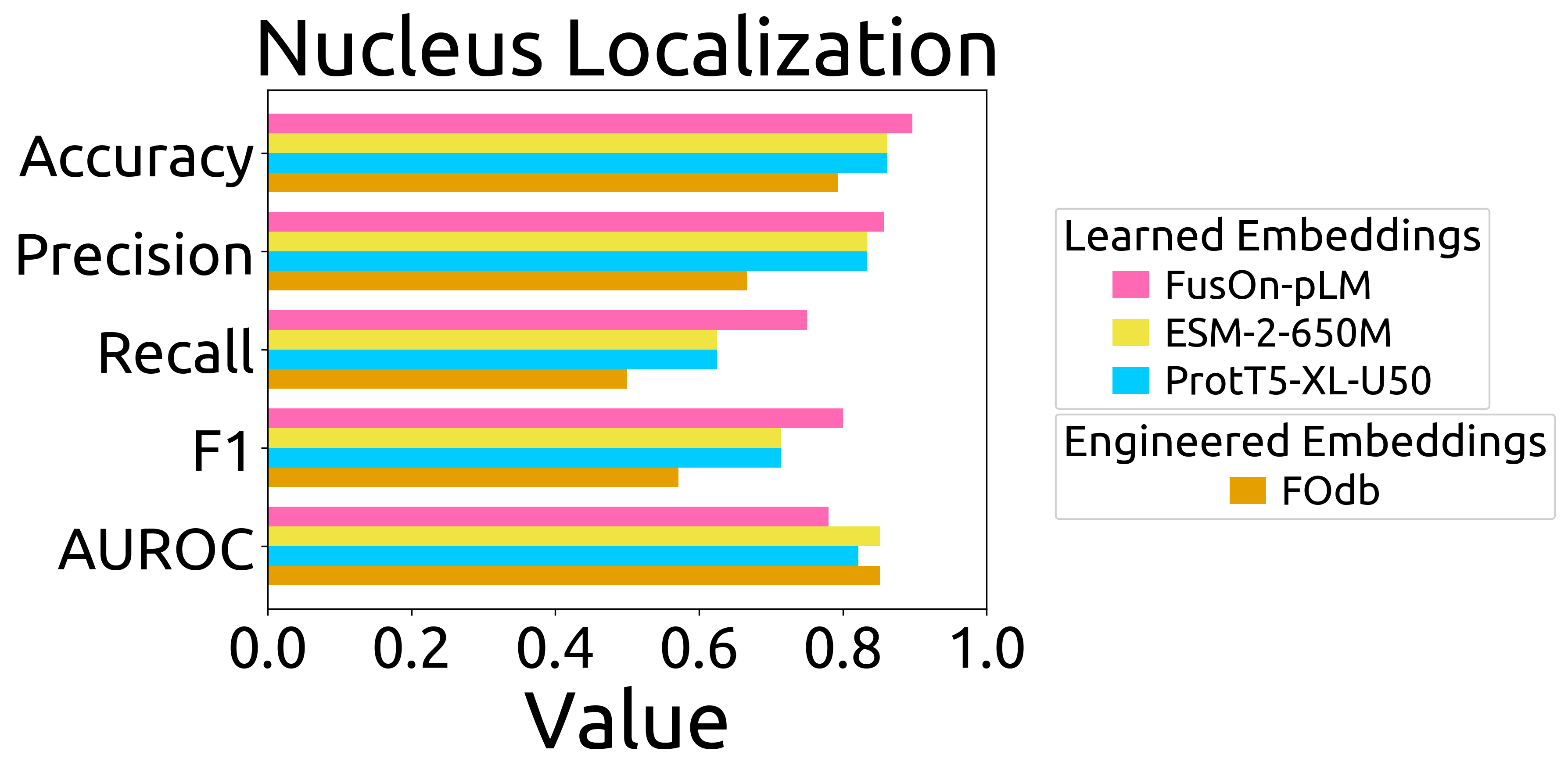
- **`nucleus_verificationFOs_barchart.png`**: bar chart of performance on the nucleus puncta prediction task (Fig. 3D), and the formatted data that went directly into the plot (`nucleus_verificationFOs_barchart_source_data.csv`)
|
| 81 |
+
|
| 82 |
+
The raw data are included in `results/final` as `cytoplasm_verificationFOs_results.csv`, `formation_verificationFOs_0.83thresh_results.csv`, and `nucleus_verificationFOs_results.csv`.
|
| 83 |
+
|
| 84 |
+
If you train a new model, the equivalents of these files will be created in `results/timestamp` for your specific configurations set in `config.py`.
|
| 85 |
+
|
| 86 |
+
To run training, enter in terminal:
|
| 87 |
+
```
|
| 88 |
+
python train.py
|
| 89 |
+
```
|
| 90 |
+
|
| 91 |
+
To regnerate plots, run
|
| 92 |
+
```
|
| 93 |
+
python plot.py
|
| 94 |
+
```
|
| 95 |
+
|
fuson_plm/benchmarking/puncta/__init__.py
ADDED
|
File without changes
|
fuson_plm/benchmarking/puncta/clean.py
ADDED
|
@@ -0,0 +1,184 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Cleans raw data to prepare FO labels and embeddings
|
| 2 |
+
from fuson_plm.utils.logging import open_logfile, log_update
|
| 3 |
+
from fuson_plm.utils.data_cleaning import find_invalid_chars
|
| 4 |
+
from fuson_plm.utils.constants import VALID_AAS
|
| 5 |
+
import pandas as pd
|
| 6 |
+
import numpy as np
|
| 7 |
+
import pickle
|
| 8 |
+
|
| 9 |
+
def find_localization(row):
|
| 10 |
+
puncta_status = row['Puncta_Status']
|
| 11 |
+
cytoplasm = (row['Cytoplasm']=='Punctate')
|
| 12 |
+
nucleus = (row['Nucleus']=='Punctate')
|
| 13 |
+
both = cytoplasm and nucleus
|
| 14 |
+
|
| 15 |
+
if puncta_status=='YES':
|
| 16 |
+
if both:
|
| 17 |
+
return 'Both'
|
| 18 |
+
else:
|
| 19 |
+
if cytoplasm:
|
| 20 |
+
return 'Cytoplasm'
|
| 21 |
+
if nucleus:
|
| 22 |
+
return 'Nucleus'
|
| 23 |
+
return np.nan
|
| 24 |
+
|
| 25 |
+
def clean_s5(df):
|
| 26 |
+
log_update("Cleaning FOdb Supplementary Table 5")
|
| 27 |
+
|
| 28 |
+
# extract only the physicochemical features used by the FO-Puncta ML model
|
| 29 |
+
retained_features = df.loc[
|
| 30 |
+
df['Low MI Set: Used In ML Model'].isin(['Yes','Yet']) # allow flexibility for typo in this DF
|
| 31 |
+
]['Parameter Label (Sup Table 2 & Matlab Scripts)'].tolist()
|
| 32 |
+
retained_features = sorted(retained_features)
|
| 33 |
+
|
| 34 |
+
# log the result
|
| 35 |
+
log_update(f'\tIsolated the {len(retained_features)} low-MI features used to train ML model')
|
| 36 |
+
for i, feat in enumerate(retained_features): log_update(f'\t\t{i+1}. {feat}')
|
| 37 |
+
|
| 38 |
+
# return the result
|
| 39 |
+
return retained_features
|
| 40 |
+
|
| 41 |
+
def make_label_df(df):
|
| 42 |
+
"""
|
| 43 |
+
Input df should be cleaned s4
|
| 44 |
+
"""
|
| 45 |
+
label_df = df[['FO_Name','AAseq','Localization','Puncta_Status','Dataset']].rename(columns={'FO_Name':'fusiongene','AAseq':'aa_seq','Dataset':'dataset'})
|
| 46 |
+
dataset_to_split_dict = {'Expressed_Set': 'train', 'Verification_Set': 'test'}
|
| 47 |
+
label_df['split'] = label_df['dataset'].apply(lambda x: dataset_to_split_dict[x])
|
| 48 |
+
label_df['nucleus'] = label_df['Localization'].apply(lambda x: 1 if x in ['Nucleus','Both'] else 0)
|
| 49 |
+
label_df['cytoplasm'] = label_df['Localization'].apply(lambda x: 1 if x in ['Cytoplasm','Both'] else 0)
|
| 50 |
+
label_df['formation'] = label_df['Puncta_Status'].apply(lambda x: 1 if x=='YES' else 0)
|
| 51 |
+
label_df = label_df[['fusiongene','aa_seq','dataset','split','nucleus','cytoplasm','formation']]
|
| 52 |
+
|
| 53 |
+
return label_df
|
| 54 |
+
|
| 55 |
+
def make_embeddings(df, physicochemical_features):
|
| 56 |
+
feat_string = '\n\t' + '\n\t'.join([str(i)+'. '+feat for i,feat in enumerate(physicochemical_features)])
|
| 57 |
+
log_update(f"\nMaking phyisochemical feature vectors.\nFeature Order: {feat_string}")
|
| 58 |
+
embeddings = {}
|
| 59 |
+
aa_seqs = df['AAseq'].unique()
|
| 60 |
+
for seq in aa_seqs:
|
| 61 |
+
feats = df.loc[df['AAseq']==seq].reset_index(drop=True)[physicochemical_features].T[0].tolist()
|
| 62 |
+
embeddings[seq] = feats
|
| 63 |
+
|
| 64 |
+
return embeddings
|
| 65 |
+
|
| 66 |
+
def clean_s4(df, retained_features):
|
| 67 |
+
log_update("Cleaning FOdb Supplementary Table 4")
|
| 68 |
+
df = df.loc[
|
| 69 |
+
df['Puncta_Status'].isin(['YES','NO'])
|
| 70 |
+
].reset_index(drop=True)
|
| 71 |
+
log_update(f'\tRemoved invalid FOs (puncta status = "Other" or "Nucleolar"). Remaining FOs: {len(df)}')
|
| 72 |
+
|
| 73 |
+
# check for duplicate sequences
|
| 74 |
+
dup_seqs = df.loc[df['AAseq'].duplicated()]['AAseq'].unique()
|
| 75 |
+
log_update(f"\tTotal duplicated sequences: {len(dup_seqs)}")
|
| 76 |
+
|
| 77 |
+
# check for invalid characters
|
| 78 |
+
df['invalid_chars'] = df['AAseq'].apply(lambda x: find_invalid_chars(x, VALID_AAS))
|
| 79 |
+
all_invalid_chars = set().union(*df['invalid_chars'])
|
| 80 |
+
log_update(f"\tChecking for invalid characters...\n\t\tFound {len(all_invalid_chars)} invalid characters")
|
| 81 |
+
for c in all_invalid_chars:
|
| 82 |
+
subset = df.loc[df['AAseq'].str.contains(c)]['AAseq'].tolist()
|
| 83 |
+
for seq in subset:
|
| 84 |
+
log_update(f"\t\tInvalid char {c} at index {seq.index(c)}/{len(seq)-1} of sequence {seq}")
|
| 85 |
+
# going to just remove the "-" from the special sequence
|
| 86 |
+
df = df.drop(columns=['invalid_chars'])
|
| 87 |
+
df.loc[
|
| 88 |
+
df['AAseq'].str.contains('-'),'AAseq'
|
| 89 |
+
] = df.loc[df['AAseq'].str.contains('-'),'AAseq'].item().replace('-','')
|
| 90 |
+
|
| 91 |
+
# change FO format to ::
|
| 92 |
+
df['FO_Name'] = df['FO_Name'].apply(lambda x: x.replace('_','::'))
|
| 93 |
+
log_update(f'\tChanged FO names to Head::Tail format')
|
| 94 |
+
|
| 95 |
+
# Isolate positive and negative sets
|
| 96 |
+
df['Localization'] = ['']*len(df)
|
| 97 |
+
df['Localization'] = df.apply(lambda row: find_localization(row), axis=1)
|
| 98 |
+
puncta_positive = df.loc[
|
| 99 |
+
df['Puncta_Status']=='YES'
|
| 100 |
+
].reset_index(drop=True)
|
| 101 |
+
puncta_negative = df.loc[
|
| 102 |
+
df['Puncta_Status']=='NO'
|
| 103 |
+
].reset_index(drop=True)
|
| 104 |
+
|
| 105 |
+
# Only keeping retained features
|
| 106 |
+
cols = list(df.columns)
|
| 107 |
+
mi_feats_included = set(retained_features).intersection(set(cols))
|
| 108 |
+
log_update(f"\tChecking for the {len(retained_features)} low-MI features... {len(mi_feats_included)} found")
|
| 109 |
+
# make sure all of these are no-na
|
| 110 |
+
for rf in retained_features:
|
| 111 |
+
# if there's NaN, log it. Make sure the only instances of np.nan are for Verification Set FOs.
|
| 112 |
+
if df[rf].isna().sum()>0:
|
| 113 |
+
nas = df.loc[df[rf].isna()]
|
| 114 |
+
log_update(f"\t\tFeature {rf} has {len(nas)} np.nan values in the following datasets:")
|
| 115 |
+
for k,v in nas['Dataset'].value_counts().items():
|
| 116 |
+
print(f'\t\t\t{k}: {v}')
|
| 117 |
+
|
| 118 |
+
df = df[['FO_Name', 'Nucleus', 'Nucleolus', 'Cytoplasm','Puncta_Status', 'Dataset', 'Localization', 'AAseq',
|
| 119 |
+
'Puncta.pred', 'Puncta.prob']+retained_features]
|
| 120 |
+
|
| 121 |
+
# Quantify localization
|
| 122 |
+
log_update(f'\n\tPuncta localization for {len(puncta_positive)} FOs where Puncta_Status==YES')
|
| 123 |
+
for k, v in puncta_positive['Localization'].value_counts().items():
|
| 124 |
+
pcnt = 100*v/sum(puncta_positive['Localization'].value_counts())
|
| 125 |
+
log_update(f'\t\t{k}: \t{v} ({pcnt:.2f}%)')
|
| 126 |
+
|
| 127 |
+
log_update("\tDataset breakdown...")
|
| 128 |
+
dataset_vc = df['Dataset'].value_counts()
|
| 129 |
+
expressed_puncta_statuses = df.loc[df['Dataset']=='Expressed_Set']['Puncta_Status'].value_counts()
|
| 130 |
+
expressed_positive_locs = puncta_positive.loc[puncta_positive['Dataset']=='Expressed_Set']['Localization'].value_counts()
|
| 131 |
+
verification_positive_locs = puncta_positive.loc[puncta_positive['Dataset']=='Verification_Set']['Localization'].value_counts()
|
| 132 |
+
verification_puncta_statuses = df.loc[df['Dataset']=='Verification_Set']['Puncta_Status'].value_counts()
|
| 133 |
+
for k, v in dataset_vc.items():
|
| 134 |
+
pcnt = 100*v/sum(dataset_vc)
|
| 135 |
+
log_update(f'\t\t{k}: \t{v} ({pcnt:.2f}%)')
|
| 136 |
+
if k=='Expressed_Set':
|
| 137 |
+
for key, val in expressed_puncta_statuses.items():
|
| 138 |
+
pcnt = 100*val/v
|
| 139 |
+
log_update(f'\t\t\t{key}: \t{val} ({pcnt:.2f}%)')
|
| 140 |
+
if key=='YES':
|
| 141 |
+
log_update('\t\t\t\tLocalizations...')
|
| 142 |
+
for key2, val2 in expressed_positive_locs.items():
|
| 143 |
+
pcnt = 100*val2/val
|
| 144 |
+
log_update(f'\t\t\t\t\t{key2}: \t{val2} ({pcnt:.2f}%)')
|
| 145 |
+
if k=='Verification_Set':
|
| 146 |
+
for key, val in verification_puncta_statuses.items():
|
| 147 |
+
pcnt = 100*val/v
|
| 148 |
+
log_update(f'\t\t\t{key}: \t{val} ({pcnt:.2f}%)')
|
| 149 |
+
if key=='YES':
|
| 150 |
+
log_update('\t\t\t\tLocalizations...')
|
| 151 |
+
for key2, val2 in verification_positive_locs.items():
|
| 152 |
+
pcnt = 100*val2/val
|
| 153 |
+
log_update(f'\t\t\t\t\t{key2}: \t{val2} ({pcnt:.2f}%)')
|
| 154 |
+
|
| 155 |
+
return df
|
| 156 |
+
|
| 157 |
+
def main():
|
| 158 |
+
LOG_PATH = 'cleaning_log.txt'
|
| 159 |
+
FODB_S4_PATH = '../../data/raw_data/FOdb_puncta.csv'
|
| 160 |
+
FODB_S5_PATH = '../../data/raw_data/FOdb_SD5.csv'
|
| 161 |
+
|
| 162 |
+
with open_logfile(LOG_PATH):
|
| 163 |
+
s4 = pd.read_csv(FODB_S4_PATH)
|
| 164 |
+
s5 = pd.read_csv(FODB_S5_PATH)
|
| 165 |
+
|
| 166 |
+
retained_features = clean_s5(s5)
|
| 167 |
+
cleaned_s4 = clean_s4(s4, retained_features)
|
| 168 |
+
|
| 169 |
+
label_df = make_label_df(cleaned_s4)
|
| 170 |
+
embeddings = make_embeddings(cleaned_s4, retained_features)
|
| 171 |
+
|
| 172 |
+
# save the results
|
| 173 |
+
cleaned_s4.to_csv('cleaned_dataset_s4.csv', index=False)
|
| 174 |
+
log_update("\nSaved cleaned table S5 to cleaned_dataset_s4.csv")
|
| 175 |
+
|
| 176 |
+
label_df.to_csv('splits.csv', index=False)
|
| 177 |
+
log_update("\nSaved train-test splits with nucleus, cytoplasm, and formation labels to splits.csv")
|
| 178 |
+
|
| 179 |
+
with open('FOdb_physicochemical_embeddings.pkl','wb') as f:
|
| 180 |
+
pickle.dump(embeddings, f)
|
| 181 |
+
log_update("\nSaved physicochemical embeddings as a dictionary to FOdb_physicochemical_embeddings.pkl")
|
| 182 |
+
|
| 183 |
+
if __name__ == '__main__':
|
| 184 |
+
main()
|
fuson_plm/benchmarking/puncta/cleaned_dataset_s4.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8f9075866f3296746c83eac61caf5c871e3a6dd54a2986896c9fd71a5a11511c
|
| 3 |
+
size 183523
|
fuson_plm/benchmarking/puncta/cleaning_log.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:22677b05ff483b17390edc30e53555097e85ce7ac6aaa3cd04aece67d3963bc1
|
| 3 |
+
size 3356
|
fuson_plm/benchmarking/puncta/config.py
ADDED
|
@@ -0,0 +1,17 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Benchmarking configs
|
| 2 |
+
BENCHMARK_FUSONPLM = True # True if you want to benchmark a FusOn-pLM Model
|
| 3 |
+
|
| 4 |
+
# FUSONPLM_CKPTS. If you've traiend your own model, this is a dictionary: key = run name, values = epochs
|
| 5 |
+
# If you want to use the trained FusOn-pLM, instead FUSONPLM_CKPTS="FusOn-pLM"
|
| 6 |
+
FUSONPLM_CKPTS= "FusOn-pLM"
|
| 7 |
+
|
| 8 |
+
# Model comparison configs
|
| 9 |
+
BENCHMARK_ESM = True # True if you want to benchmark ESM-2-650M
|
| 10 |
+
BENCHMARK_PROTT5 = True # True if you want to benchmark ProtT5
|
| 11 |
+
BENCHMARK_FO_PUNCTA_ML = True # True if you want to benchmark FO-Puncta-ML from the FOdb paper
|
| 12 |
+
|
| 13 |
+
# Overwriting configs
|
| 14 |
+
PERMISSION_TO_OVERWRITE = False # if False, script will halt if it believes these embeddings have already been made.
|
| 15 |
+
|
| 16 |
+
# GPU configs
|
| 17 |
+
CUDA_VISIBLE_DEVICES="0" # GPUs to make visible for this process
|
fuson_plm/benchmarking/puncta/plot.py
ADDED
|
@@ -0,0 +1,244 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import matplotlib.pyplot as plt
|
| 2 |
+
import matplotlib.patches as mpatches
|
| 3 |
+
import seaborn as sns
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import numpy as np
|
| 6 |
+
import os
|
| 7 |
+
import matplotlib.colors as mcolors
|
| 8 |
+
from fuson_plm.utils.visualizing import set_font
|
| 9 |
+
|
| 10 |
+
fo_puncta_db_training_thresh31 = pd.DataFrame(data={
|
| 11 |
+
'Model Type': ['fo_puncta_ml'],
|
| 12 |
+
'Model Name': ['fo_puncta_ml_literature'],
|
| 13 |
+
'Model Epoch': np.nan,
|
| 14 |
+
'Accuracy': 0.81,
|
| 15 |
+
'Precision': 0.78,
|
| 16 |
+
'Recall': 0.98,
|
| 17 |
+
'F1 Score': 0.87,
|
| 18 |
+
'AUROC': 0.88,
|
| 19 |
+
'AUPRC': 0.94
|
| 20 |
+
})
|
| 21 |
+
|
| 22 |
+
fo_puncta_db_verification_thresh83 = pd.DataFrame(data={
|
| 23 |
+
'Model Type': ['fo_puncta_ml'],
|
| 24 |
+
'Model Name': ['fo_puncta_ml_literature'],
|
| 25 |
+
'Model Epoch': np.nan,
|
| 26 |
+
'Accuracy': 0.79,
|
| 27 |
+
'Precision': 0.81,
|
| 28 |
+
'Recall': 0.89,
|
| 29 |
+
'F1 Score': 0.85,
|
| 30 |
+
'AUROC': 0.73,
|
| 31 |
+
'AUPRC': 0.82
|
| 32 |
+
})
|
| 33 |
+
|
| 34 |
+
# Method for lengthening the model name
|
| 35 |
+
def lengthen_model_name(row):
|
| 36 |
+
name = row['Model Name']
|
| 37 |
+
epoch = row['Model Epoch']
|
| 38 |
+
|
| 39 |
+
if 'esm' in name:
|
| 40 |
+
return name
|
| 41 |
+
if 'puncta' in name:
|
| 42 |
+
return name
|
| 43 |
+
|
| 44 |
+
return f'{name}_e{epoch}'
|
| 45 |
+
|
| 46 |
+
# Method for shortening the model name for display
|
| 47 |
+
def shorten_model_name(row):
|
| 48 |
+
name = row['Model Name']
|
| 49 |
+
epoch = row['Model Epoch']
|
| 50 |
+
|
| 51 |
+
if 'esm' in name:
|
| 52 |
+
return 'ESM-2-650M'
|
| 53 |
+
if name=='fo_puncta_ml':
|
| 54 |
+
return 'FO-Puncta-ML'
|
| 55 |
+
if name=='fo_puncta_ml_literature':
|
| 56 |
+
return 'FO-Puncta-ML Lit'
|
| 57 |
+
if name=="prot_t5_xl_half_uniref50_enc":
|
| 58 |
+
return 'ProtT5-XL-U50' # this is waht they call it in the paper
|
| 59 |
+
|
| 60 |
+
if 'snp_' in name:
|
| 61 |
+
prob_type = 'snp'
|
| 62 |
+
elif 'uniform_' in name:
|
| 63 |
+
prob_type = 'uni'
|
| 64 |
+
|
| 65 |
+
layers = name.split('layers')[0].split('_')[-1]
|
| 66 |
+
dt = name.split('mask')[1].split('-', 1)[1]
|
| 67 |
+
|
| 68 |
+
return f'{prob_type}_{layers}L_{dt}_e{epoch}'
|
| 69 |
+
|
| 70 |
+
def make_final_bar(dataframe, title, save_path):
|
| 71 |
+
set_font()
|
| 72 |
+
df = dataframe.copy(deep=True)
|
| 73 |
+
|
| 74 |
+
# Pivot the DataFrame to have metrics as rows and names as columns, and reorder columns
|
| 75 |
+
pivot_df = df.pivot(index='Metric', columns='Name', values='Value')
|
| 76 |
+
ordered_columns = [x for x in ['FOdb','ProtT5-XL-U50', 'ESM-2-650M', 'FusOn-pLM'] if x in pivot_df.columns]
|
| 77 |
+
pivot_df = pivot_df[ordered_columns]
|
| 78 |
+
|
| 79 |
+
# Define the groups
|
| 80 |
+
engineered_embeddings = ['FOdb']
|
| 81 |
+
deep_learning_embeddings = ['ProtT5-XL-U50', 'ESM-2-650M', 'FusOn-pLM']
|
| 82 |
+
|
| 83 |
+
# Reorder the metrics
|
| 84 |
+
metric_order = ['Accuracy', 'Precision', 'Recall', 'F1', 'AUROC'][::-1]
|
| 85 |
+
pivot_df = pivot_df.reindex(metric_order)
|
| 86 |
+
|
| 87 |
+
# Plotting
|
| 88 |
+
fig, ax = plt.subplots(figsize=(8, 6), dpi=300) # Increased figure size for better legend placement
|
| 89 |
+
|
| 90 |
+
# Define bar width and positions
|
| 91 |
+
bar_width = 0.2
|
| 92 |
+
indices = np.arange(len(pivot_df))
|
| 93 |
+
|
| 94 |
+
# Use a colorblind-friendly color scheme from tableau
|
| 95 |
+
color_map = {
|
| 96 |
+
#'One-Hot': "#999999",
|
| 97 |
+
'FOdb': "#E69F00",
|
| 98 |
+
'ESM-2-650M': "#F0E442",
|
| 99 |
+
'FusOn-pLM': "#FF69B4",
|
| 100 |
+
'ProtT5-XL-U50': "#00ccff" # light blue
|
| 101 |
+
}
|
| 102 |
+
colors = [color_map[col] for col in ordered_columns]
|
| 103 |
+
|
| 104 |
+
# Plot bars for each category and add them to appropriate legend groups
|
| 105 |
+
engineered_handles = []
|
| 106 |
+
deep_learning_handles = []
|
| 107 |
+
for i, (name, color) in enumerate(zip(pivot_df.columns, colors)):
|
| 108 |
+
bars = ax.barh(indices + i * bar_width, pivot_df[name], bar_width, label=name, color=color)
|
| 109 |
+
if name in engineered_embeddings:
|
| 110 |
+
engineered_handles.append(bars[0])
|
| 111 |
+
else:
|
| 112 |
+
deep_learning_handles.append(bars[0])
|
| 113 |
+
|
| 114 |
+
# Add bold black asterisks next to the winning bars for each category (could be multiple)
|
| 115 |
+
#for j, metric in enumerate(pivot_df.index):
|
| 116 |
+
# max_value = pivot_df.loc[metric].max()
|
| 117 |
+
# max_indices = pivot_df.loc[metric][pivot_df.loc[metric] == max_value].index
|
| 118 |
+
# for max_name in max_indices:
|
| 119 |
+
# max_index = list(pivot_df.columns).index(max_name)
|
| 120 |
+
# ax.text(max_value + 0.01, j + max_index * bar_width - bar_width / 4, '*',
|
| 121 |
+
# color='black', fontsize=12, fontweight='bold', ha='center', va='center')
|
| 122 |
+
|
| 123 |
+
# Set labels, ticks, and title
|
| 124 |
+
plt.xlabel('Value', fontsize=44) # Adjusted font size
|
| 125 |
+
ax.set_yticks(indices + bar_width * 1.5)
|
| 126 |
+
ax.set_xlim([0, 1])
|
| 127 |
+
ax.set_yticklabels(pivot_df.index)
|
| 128 |
+
# make the xticklabels size 24
|
| 129 |
+
ax.tick_params(axis='x')
|
| 130 |
+
ax.set_title(title, fontsize=44) # Adjusted font size
|
| 131 |
+
|
| 132 |
+
# Setting font size for tick labels
|
| 133 |
+
for label in plt.gca().get_xticklabels():
|
| 134 |
+
label.set_fontsize(32) # Adjusted font size
|
| 135 |
+
for label in plt.gca().get_yticklabels():
|
| 136 |
+
label.set_fontsize(32) # Adjusted font size
|
| 137 |
+
|
| 138 |
+
# Create two separate legends
|
| 139 |
+
if engineered_handles:
|
| 140 |
+
legend1 = fig.legend(
|
| 141 |
+
engineered_handles[::-1],
|
| 142 |
+
[emb for emb in engineered_embeddings if emb in ordered_columns][::-1],
|
| 143 |
+
loc='center left',
|
| 144 |
+
bbox_to_anchor=(1, 0.4),
|
| 145 |
+
title="Engineered Embeddings",
|
| 146 |
+
title_fontsize=24) # Adjusted font size
|
| 147 |
+
if deep_learning_handles:
|
| 148 |
+
legend2 = fig.legend(
|
| 149 |
+
deep_learning_handles[::-1],
|
| 150 |
+
[emb for emb in deep_learning_embeddings if emb in ordered_columns][::-1],
|
| 151 |
+
loc='center left',
|
| 152 |
+
bbox_to_anchor=(1, 0.6),
|
| 153 |
+
title="Learned Embeddings",
|
| 154 |
+
title_fontsize=24) # Adjusted font size
|
| 155 |
+
|
| 156 |
+
# Adjust legend text size
|
| 157 |
+
if engineered_handles:
|
| 158 |
+
ax.add_artist(legend1)
|
| 159 |
+
for text in legend1.get_texts():
|
| 160 |
+
text.set_fontsize(22) # Adjusted font size
|
| 161 |
+
for handle in legend1.legendHandles:
|
| 162 |
+
if isinstance(handle, mpatches.Patch):
|
| 163 |
+
handle.set_height(15) # Adjust height
|
| 164 |
+
handle.set_width(20) # Adjust width
|
| 165 |
+
elif hasattr(handle, '_sizes'):
|
| 166 |
+
handle._sizes = [200] # Increase marker size in the legend
|
| 167 |
+
|
| 168 |
+
if deep_learning_handles:
|
| 169 |
+
ax.add_artist(legend2)
|
| 170 |
+
for text in legend2.get_texts():
|
| 171 |
+
text.set_fontsize(22) # Adjusted font size
|
| 172 |
+
for handle in legend2.legendHandles:
|
| 173 |
+
if isinstance(handle, mpatches.Patch):
|
| 174 |
+
handle.set_height(15) # Adjust height
|
| 175 |
+
handle.set_width(20) # Adjust width
|
| 176 |
+
elif hasattr(handle, '_sizes'):
|
| 177 |
+
handle._sizes = [200] # Increase marker size in the legend
|
| 178 |
+
|
| 179 |
+
plt.tight_layout() # Adjust layout to make room for the legends
|
| 180 |
+
|
| 181 |
+
# Save the plot to a file
|
| 182 |
+
plt.savefig(save_path, dpi=300, bbox_inches='tight')
|
| 183 |
+
|
| 184 |
+
plt.show()
|
| 185 |
+
|
| 186 |
+
def prepare_data_for_bar(results_dir, task, split, thresh=None):
|
| 187 |
+
fname = f"{task}_{split}FOs_results.csv"
|
| 188 |
+
if thresh is not None: fname = f"{task}_{split}FOs_{thresh}thresh_results.csv"
|
| 189 |
+
image_save_path = results_dir + '/figures/' + fname.split('_results.csv')[0]+'_barchart.png'
|
| 190 |
+
|
| 191 |
+
data = pd.read_csv(f"{results_dir}/{fname}")
|
| 192 |
+
data = data.loc[
|
| 193 |
+
data['Model Name'].isin(['best',
|
| 194 |
+
'fo_puncta_ml',
|
| 195 |
+
'esm2_t33_650M_UR50D',
|
| 196 |
+
'prot_t5_xl_half_uniref50_enc'])
|
| 197 |
+
]
|
| 198 |
+
data = pd.DataFrame(data = {
|
| 199 |
+
'Name': data['Model Name'].tolist() * 5,
|
| 200 |
+
'Metric': ['Accuracy', 'Accuracy', 'Accuracy','Accuracy',
|
| 201 |
+
'Precision', 'Precision', 'Precision', 'Precision',
|
| 202 |
+
'Recall', 'Recall', 'Recall', 'Recall',
|
| 203 |
+
'F1', 'F1', 'F1','F1',
|
| 204 |
+
'AUROC', 'AUROC', 'AUROC','AUROC'],
|
| 205 |
+
'Value': data['Accuracy'].tolist() + data['Precision'].tolist() + data['Recall'].tolist() + data['F1 Score'].tolist() + data['AUROC'].tolist()
|
| 206 |
+
}
|
| 207 |
+
)
|
| 208 |
+
rename_dict = {'fo_puncta_ml': 'FOdb',
|
| 209 |
+
'esm2_t33_650M_UR50D':'ESM-2-650M',
|
| 210 |
+
'best':'FusOn-pLM',
|
| 211 |
+
'prot_t5_xl_half_uniref50_enc': 'ProtT5-XL-U50'}
|
| 212 |
+
data['Name'] = data['Name'].map(rename_dict)
|
| 213 |
+
return data, image_save_path
|
| 214 |
+
|
| 215 |
+
def make_all_final_bar_charts(results_dir):
|
| 216 |
+
# Puncta verification
|
| 217 |
+
data, image_save_path = prepare_data_for_bar(results_dir,"formation","verification",thresh=0.83)
|
| 218 |
+
data_cp = data.copy(deep=True)
|
| 219 |
+
data_cp["Value"] = data_cp["Value"].round(3)
|
| 220 |
+
data_cp.to_csv(image_save_path.replace(".png","_source_data.csv"),index=False)
|
| 221 |
+
make_final_bar(data, "Puncta Propensity", image_save_path)
|
| 222 |
+
|
| 223 |
+
# Nucleus verification
|
| 224 |
+
data, image_save_path = prepare_data_for_bar(results_dir,"nucleus","verification",thresh=None)
|
| 225 |
+
data_cp = data.copy(deep=True)
|
| 226 |
+
data_cp["Value"] = data_cp["Value"].round(3)
|
| 227 |
+
data_cp.to_csv(image_save_path.replace(".png","_source_data.csv"),index=False)
|
| 228 |
+
make_final_bar(data, "Nucleus Localization", image_save_path)
|
| 229 |
+
|
| 230 |
+
# Cytoplasm verification
|
| 231 |
+
data, image_save_path = prepare_data_for_bar(results_dir,"cytoplasm","verification",thresh=None)
|
| 232 |
+
data_cp = data.copy(deep=True)
|
| 233 |
+
data_cp["Value"] = data_cp["Value"].round(3)
|
| 234 |
+
data_cp.to_csv(image_save_path.replace(".png","_source_data.csv"),index=False)
|
| 235 |
+
make_final_bar(data, "Cytoplasm Localization", image_save_path)
|
| 236 |
+
|
| 237 |
+
def main():
|
| 238 |
+
# Read in the input data
|
| 239 |
+
results_dir="results/final"
|
| 240 |
+
os.makedirs(f"{results_dir}/figures",exist_ok=True)
|
| 241 |
+
make_all_final_bar_charts(results_dir)
|
| 242 |
+
|
| 243 |
+
if __name__ == '__main__':
|
| 244 |
+
main()
|
fuson_plm/benchmarking/puncta/results/final/cytoplasm_verificationFOs_results.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:800f935f72b089b357fb4b0ac22a4c75a09a4578e44fac2c20a297c60c76df76
|
| 3 |
+
size 871
|
fuson_plm/benchmarking/puncta/results/final/figures/cytoplasm_verificationFOs_barchart.png
ADDED
|
fuson_plm/benchmarking/puncta/results/final/figures/cytoplasm_verificationFOs_barchart_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:06aa241a68bff40ae38cd6d484c4ff3ebf4d8613fb0e671576a3f07b6977dbda
|
| 3 |
+
size 470
|
fuson_plm/benchmarking/puncta/results/final/figures/formation_verificationFOs_0.83thresh_barchart.png
ADDED

|
fuson_plm/benchmarking/puncta/results/final/figures/formation_verificationFOs_0.83thresh_barchart_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:85ad0497edcb0438fafe20d2807afb694114bdc3a73401ca0ed6b739baca1603
|
| 3 |
+
size 472
|
fuson_plm/benchmarking/puncta/results/final/figures/nucleus_verificationFOs_barchart.png
ADDED
|
fuson_plm/benchmarking/puncta/results/final/figures/nucleus_verificationFOs_barchart_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:73697291e6f1d8036fd089babbde87e39c30d040e98a2c20d71dfb202925e316
|
| 3 |
+
size 472
|
fuson_plm/benchmarking/puncta/results/final/formation_verificationFOs_0.83thresh_results.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:72c68d45ca772a2bded7473803767c12dbafa4bac09bc10aed70a075c386682c
|
| 3 |
+
size 888
|
fuson_plm/benchmarking/puncta/results/final/nucleus_verificationFOs_results.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:37d6fc7ec393c48756c286e01ddb942b8b98b03564f22a099d01e2bd537f33ca
|
| 3 |
+
size 887
|
fuson_plm/benchmarking/puncta/splits.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:44f627efa4f76a35b2a4a83be77ae8815c7b728e3ca2ca5127d8177789127f7e
|
| 3 |
+
size 133807
|
fuson_plm/benchmarking/puncta/train.py
ADDED
|
@@ -0,0 +1,155 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import time
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import numpy as np
|
| 5 |
+
import pickle
|
| 6 |
+
import os
|
| 7 |
+
|
| 8 |
+
from fuson_plm.benchmarking.xgboost_predictor import train_final_predictor, evaluate_predictor, train_predictor_xval
|
| 9 |
+
from fuson_plm.benchmarking.embed import embed_dataset_for_benchmark
|
| 10 |
+
import fuson_plm.benchmarking.puncta.config as config
|
| 11 |
+
from fuson_plm.benchmarking.puncta.plot import make_all_final_bar_charts
|
| 12 |
+
from fuson_plm.utils.logging import log_update, open_logfile, print_configpy, get_local_time, CustomParams
|
| 13 |
+
|
| 14 |
+
def check_splits(df):
|
| 15 |
+
# make sure everything has a split
|
| 16 |
+
if len(df.loc[df['split'].isna()])>0:
|
| 17 |
+
raise Exception("Error: not every benchmarking sequence has been allocated to a split (train or test)")
|
| 18 |
+
# make sure the only things are train and test
|
| 19 |
+
if len({'train','test'} - set(df['split'].unique()))!=0:
|
| 20 |
+
raise Exception("Error: splits column should only have \'train\' and \'test\'.")
|
| 21 |
+
# make sure there are no duplicate sequences
|
| 22 |
+
if len(df.loc[df['aa_seq'].duplicated()])>0:
|
| 23 |
+
raise Exception("Error: duplicate sequences provided")
|
| 24 |
+
|
| 25 |
+
def train_and_evaluate_puncta_predictor(details, splits_with_embeddings,outdir,task='nucleus',class1_thresh=0.5,n_estimators=50,tree_method="hist"):
|
| 26 |
+
"""
|
| 27 |
+
task = 'nucleus', 'cytoplasm', or 'formation'
|
| 28 |
+
"""
|
| 29 |
+
# unpack the details dictioanry
|
| 30 |
+
benchmark_model_type = details['model_type']
|
| 31 |
+
benchmark_model_name = details['model']
|
| 32 |
+
benchmark_model_epoch = details['epoch']
|
| 33 |
+
|
| 34 |
+
# prepare train and test sets for model
|
| 35 |
+
train_split = splits_with_embeddings.loc[splits_with_embeddings['split']=='train'].reset_index(drop=True)
|
| 36 |
+
test_split = splits_with_embeddings.loc[splits_with_embeddings['split']=='test'].reset_index(drop=True)
|
| 37 |
+
|
| 38 |
+
X_train = np.array(train_split['embedding'].tolist())
|
| 39 |
+
y_train = np.array(train_split[task].tolist())
|
| 40 |
+
X_test = np.array(test_split['embedding'].tolist())
|
| 41 |
+
y_test = np.array(test_split[task].tolist())
|
| 42 |
+
|
| 43 |
+
# Train the final model on all the data
|
| 44 |
+
clf = train_final_predictor(X_train, y_train, n_estimators=n_estimators, tree_method=tree_method)
|
| 45 |
+
|
| 46 |
+
# Evaluate it
|
| 47 |
+
automatic_stats_df, custom_stats_df = evaluate_predictor(clf, X_test, y_test, class1_thresh=class1_thresh)
|
| 48 |
+
|
| 49 |
+
# Add the model details back in
|
| 50 |
+
cols = list(automatic_stats_df.columns)
|
| 51 |
+
automatic_stats_df['Model Type'] = [benchmark_model_type]
|
| 52 |
+
automatic_stats_df['Model Name'] = [benchmark_model_name]
|
| 53 |
+
automatic_stats_df['Model Epoch'] = [benchmark_model_epoch]
|
| 54 |
+
newcols = ['Model Type','Model Name','Model Epoch'] + cols
|
| 55 |
+
automatic_stats_df = automatic_stats_df[newcols]
|
| 56 |
+
|
| 57 |
+
cols = list(custom_stats_df.columns)
|
| 58 |
+
custom_stats_df['Model Type'] = [benchmark_model_type]
|
| 59 |
+
custom_stats_df['Model Name'] = [benchmark_model_name]
|
| 60 |
+
custom_stats_df['Model Epoch'] = [benchmark_model_epoch]
|
| 61 |
+
newcols = ['Model Type','Model Name','Model Epoch'] + cols
|
| 62 |
+
custom_stats_df = custom_stats_df[newcols]
|
| 63 |
+
|
| 64 |
+
# Save automatic results (for nucleus and cytoplasm)
|
| 65 |
+
if task!="formation":
|
| 66 |
+
automatic_stats_path = f'{outdir}/{task}_verificationFOs_results.csv'
|
| 67 |
+
if not(os.path.exists(automatic_stats_path)):
|
| 68 |
+
automatic_stats_df.to_csv(automatic_stats_path,index=False)
|
| 69 |
+
else:
|
| 70 |
+
automatic_stats_df.to_csv(automatic_stats_path,mode='a',index=False,header=False)
|
| 71 |
+
|
| 72 |
+
# Save custom threshold results (only if it's formation)
|
| 73 |
+
if task=="formation":
|
| 74 |
+
custom_stats_path = f'{outdir}/{task}_verificationFOs_{class1_thresh}thresh_results.csv'
|
| 75 |
+
if not(os.path.exists(custom_stats_path)):
|
| 76 |
+
custom_stats_df.to_csv(custom_stats_path,index=False)
|
| 77 |
+
else:
|
| 78 |
+
custom_stats_df.to_csv(custom_stats_path,mode='a',index=False,header=False)
|
| 79 |
+
|
| 80 |
+
def main():
|
| 81 |
+
# make output directory for this run
|
| 82 |
+
os.makedirs('results',exist_ok=True)
|
| 83 |
+
output_dir = f'results/{get_local_time()}'
|
| 84 |
+
os.makedirs(output_dir,exist_ok=True)
|
| 85 |
+
|
| 86 |
+
with open_logfile(f'{output_dir}/puncta_benchmark_log.txt'):
|
| 87 |
+
# print configurations
|
| 88 |
+
print_configpy(config)
|
| 89 |
+
|
| 90 |
+
# Verify that the environment variables are set correctly
|
| 91 |
+
os.environ['CUDA_VISIBLE_DEVICES'] = config.CUDA_VISIBLE_DEVICES
|
| 92 |
+
log_update("\nChecking on environment variables...")
|
| 93 |
+
log_update(f"\tCUDA_VISIBLE_DEVICES: {os.environ.get('CUDA_VISIBLE_DEVICES')}")
|
| 94 |
+
|
| 95 |
+
# make embeddings if needed
|
| 96 |
+
all_embedding_paths = embed_dataset_for_benchmark(
|
| 97 |
+
fuson_ckpts=config.FUSONPLM_CKPTS,
|
| 98 |
+
input_data_path='splits.csv', input_fname='FOdb_puncta_sequences',
|
| 99 |
+
average=True, seq_col='aa_seq',
|
| 100 |
+
benchmark_fusonplm=config.BENCHMARK_FUSONPLM,
|
| 101 |
+
benchmark_esm=config.BENCHMARK_ESM,
|
| 102 |
+
benchmark_fo_puncta_ml=config.BENCHMARK_FO_PUNCTA_ML,
|
| 103 |
+
benchmark_prott5 = config.BENCHMARK_PROTT5,
|
| 104 |
+
overwrite=config.PERMISSION_TO_OVERWRITE)
|
| 105 |
+
|
| 106 |
+
# load the splits with labels
|
| 107 |
+
splits = pd.read_csv('splits.csv')
|
| 108 |
+
# perform some sanity checks on the splits
|
| 109 |
+
check_splits(splits)
|
| 110 |
+
n_train = len(splits.loc[splits['split']=='train'])
|
| 111 |
+
n_test = len(splits.loc[splits['split']=='test'])
|
| 112 |
+
log_update(f"\nSplit breakdown...\n\t{n_train} Training FOs\n\t{n_test} Verification FOs")
|
| 113 |
+
|
| 114 |
+
# set training constants
|
| 115 |
+
train_params = CustomParams(
|
| 116 |
+
N_ESTIMATORS = 50,
|
| 117 |
+
TREE_METHOD = "hist",
|
| 118 |
+
CLASS1_THRESHOLDS = {
|
| 119 |
+
'nucleus': 0.83,
|
| 120 |
+
'cytoplasm': 0.83,
|
| 121 |
+
'formation': 0.83
|
| 122 |
+
},
|
| 123 |
+
)
|
| 124 |
+
log_update("\nTraining configs:")
|
| 125 |
+
train_params.print_config(indent='\t')
|
| 126 |
+
|
| 127 |
+
log_update("\nTraining models")
|
| 128 |
+
# loop through the embedding paths and train each one
|
| 129 |
+
for embedding_path, details in all_embedding_paths.items():
|
| 130 |
+
log_update(f"\tBenchmarking embeddings at: {embedding_path}")
|
| 131 |
+
try:
|
| 132 |
+
with open(embedding_path, "rb") as f:
|
| 133 |
+
embeddings = pickle.load(f)
|
| 134 |
+
except:
|
| 135 |
+
raise Exception(f"Cannot read embeddings from {embedding_path}")
|
| 136 |
+
|
| 137 |
+
# combine the embeddings and splits into one dataframe
|
| 138 |
+
splits_with_embeddings = pd.DataFrame.from_dict(embeddings.items())
|
| 139 |
+
splits_with_embeddings = splits_with_embeddings.rename(columns={0: 'aa_seq', 1: 'embedding'})
|
| 140 |
+
splits_with_embeddings = pd.merge(splits_with_embeddings, splits, on='aa_seq',how='left')
|
| 141 |
+
|
| 142 |
+
for task in ['nucleus','cytoplasm','formation']:
|
| 143 |
+
log_update(f"\t\tTask: {task}")
|
| 144 |
+
train_and_evaluate_puncta_predictor(details, splits_with_embeddings, output_dir, task=task,
|
| 145 |
+
class1_thresh=train_params.CLASS1_THRESHOLDS[task],
|
| 146 |
+
n_estimators=train_params.N_ESTIMATORS,tree_method=train_params.TREE_METHOD)
|
| 147 |
+
|
| 148 |
+
log_update(f"\nMaking summary figures:\n")
|
| 149 |
+
log_update(f"\tbar charts...")
|
| 150 |
+
os.makedirs(f"{output_dir}/figures",exist_ok=True)
|
| 151 |
+
make_all_final_bar_charts(output_dir)
|
| 152 |
+
log_update(f"\tDone.")
|
| 153 |
+
|
| 154 |
+
if __name__ == '__main__':
|
| 155 |
+
main()
|