data cleaning, blast, and splitting code with source data, also deleting unnecessary files
Browse files- fuson_plm/data/README.md +7 -2
- fuson_plm/data/blast/README.md +2 -1
- model/model.pth → fuson_plm/data/blast/figures/identities_hist_source_data.csv +2 -2
- fuson_plm/data/blast/plot.py +7 -1
- fuson_plm/data/split_vis.py +14 -307
- fuson_plm/data/splits/combined_plot.png +0 -0
- fuson_plm/data/splits/split_vis/aa_comp.png +0 -0
- fuson_plm/data/splits/split_vis/aa_comp_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/combined_plot.png +0 -0
- fuson_plm/data/splits/split_vis/length_distributions.png +0 -0
- fuson_plm/data/splits/split_vis/scatterplot.png +0 -0
- fuson_plm/data/splits/split_vis/scatterplot_benchmark_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/scatterplot_test_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/scatterplot_train_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/scatterplot_val_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/shannon_entropy_plot.png +0 -0
- fuson_plm/data/splits/split_vis/shannon_entropy_plot_test_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/shannon_entropy_plot_train_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/shannon_entropy_plot_val_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/test_lengths_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/train_lengths_source_data.csv +3 -0
- fuson_plm/data/splits/split_vis/val_lengths_source_data.csv +3 -0
- fuson_plm/utils/visualizing.py +87 -37
fuson_plm/data/README.md
CHANGED
|
@@ -41,7 +41,7 @@ data/
|
|
| 41 |
- **`cluster.py`**: script for clustering the processed data in fuson_db.csv. Print statements in this code produce `clustering_log.txt`.
|
| 42 |
- **`config.py`**: configs for the cleaning, clustering, and splitting scripts.
|
| 43 |
- **`split.py`**: script for splitting the data, post-clusteirng. Print statements in this code produce `splitting_log.txt`.
|
| 44 |
-
- **`split_vis.py`** script with code for the plots in `splits/combined_plot.png`, which describe the content of the train, validation, and test splits (length distribution, Shannon Entropy, amino acid frequencies, and cluster sizes)
|
| 45 |
|
| 46 |
#### Usage
|
| 47 |
To repeat our cleaning, clustering, and splitting process, proceed as follows.
|
|
@@ -85,7 +85,12 @@ python split.py
|
|
| 85 |
This script will create the following files:
|
| 86 |
- **`splits/train_cluster_split.csv`, `splits/val_cluster_split.csv`, `splits/test_cluster_split.csv`**: The subsets of `clustering/mmseqs_full_results.csv` that have been partitioned into the train, validation, and test sets respectively.
|
| 87 |
- **`splits/train_df.csv`, `splits/val_df.csv`, `splits/test_df.csv`**: The train, validation, and testing splits used to train FusOn-pLM. Columns: `sequence`,`member length`
|
| 88 |
-
- **`
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 89 |
|
| 90 |
### BLAST
|
| 91 |
We ran BLAST to get the best alignment of each sequence in FusOn-DB to a protein in SwissProt. See the README in the `blast` folder for more details.
|
|
|
|
| 41 |
- **`cluster.py`**: script for clustering the processed data in fuson_db.csv. Print statements in this code produce `clustering_log.txt`.
|
| 42 |
- **`config.py`**: configs for the cleaning, clustering, and splitting scripts.
|
| 43 |
- **`split.py`**: script for splitting the data, post-clusteirng. Print statements in this code produce `splitting_log.txt`.
|
| 44 |
+
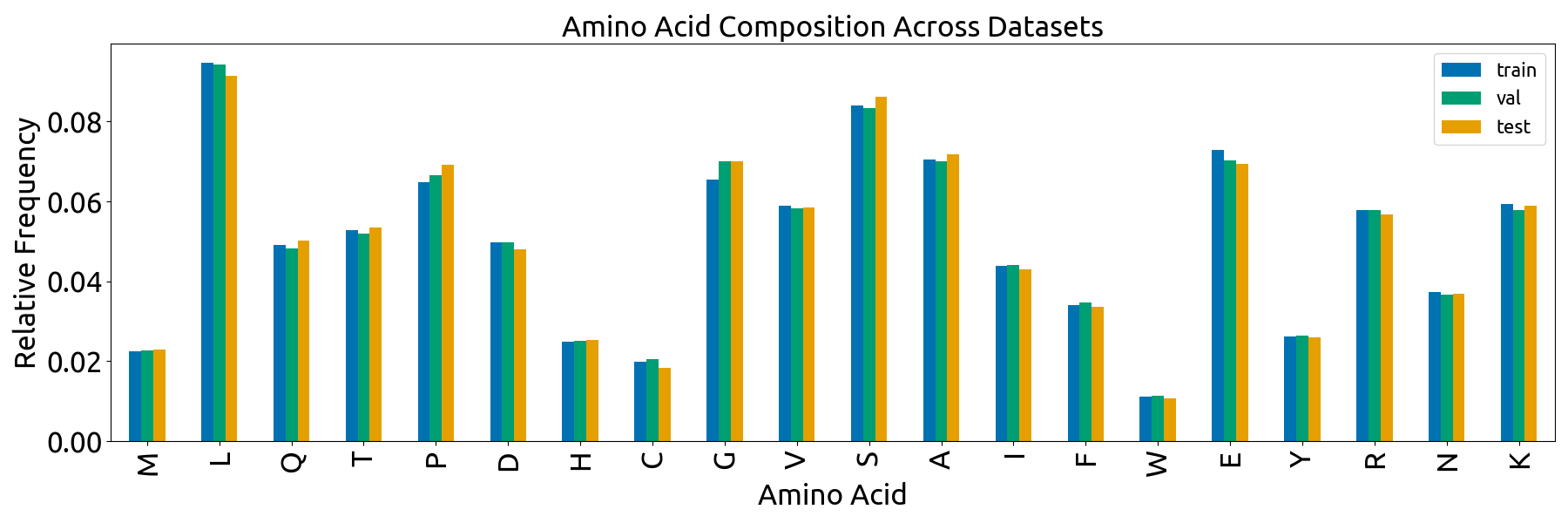
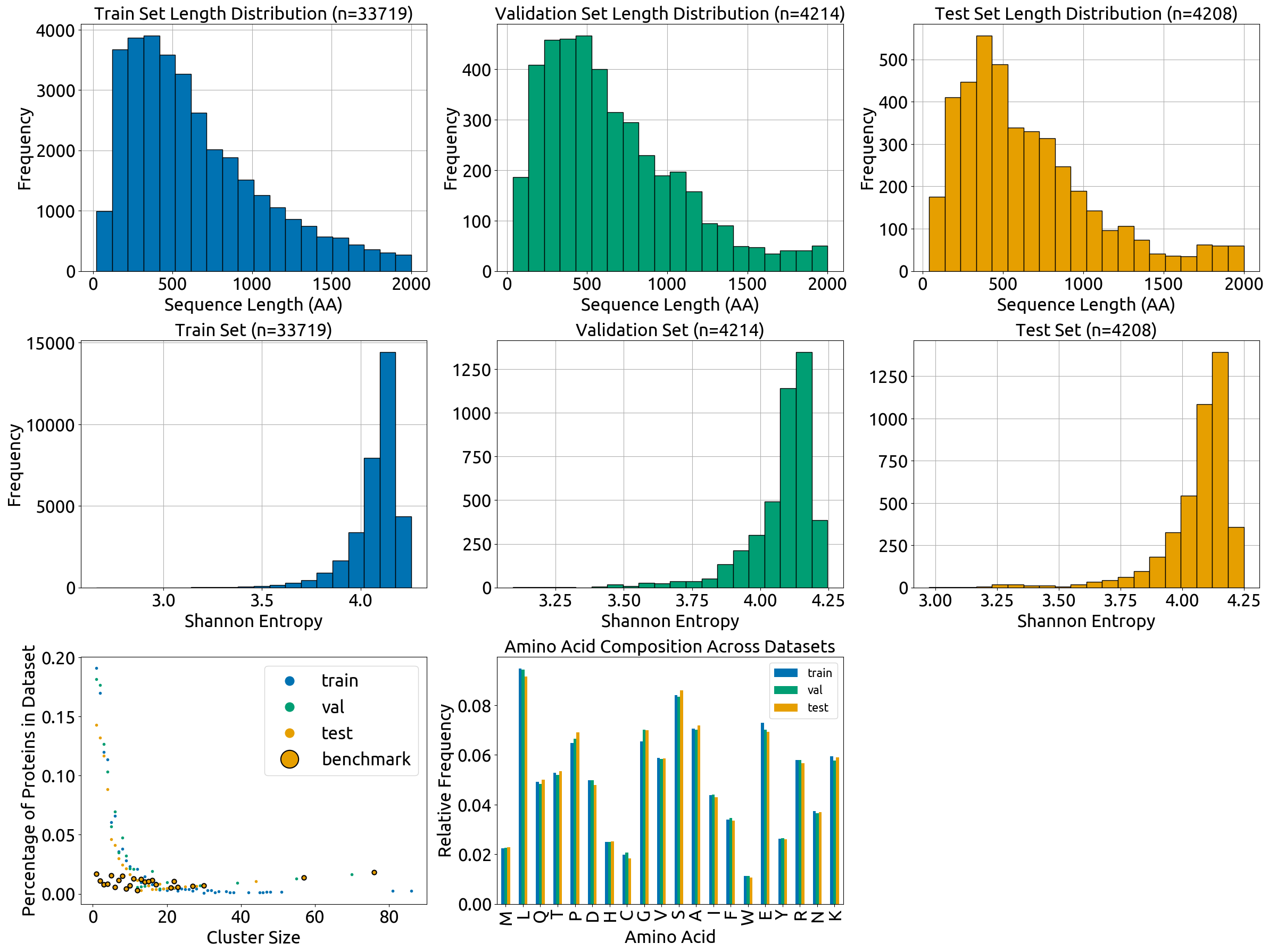
- **`split_vis.py`** script with code for the plots in `splits/combined_plot.png`, which describe the content of the train, validation, and test splits (length distribution, Shannon Entropy, amino acid frequencies, and cluster sizes). Note that many of the methods are defined in `fuson_plm/utils/visualizing.py`.
|
| 45 |
|
| 46 |
#### Usage
|
| 47 |
To repeat our cleaning, clustering, and splitting process, proceed as follows.
|
|
|
|
| 85 |
This script will create the following files:
|
| 86 |
- **`splits/train_cluster_split.csv`, `splits/val_cluster_split.csv`, `splits/test_cluster_split.csv`**: The subsets of `clustering/mmseqs_full_results.csv` that have been partitioned into the train, validation, and test sets respectively.
|
| 87 |
- **`splits/train_df.csv`, `splits/val_df.csv`, `splits/test_df.csv`**: The train, validation, and testing splits used to train FusOn-pLM. Columns: `sequence`,`member length`
|
| 88 |
+
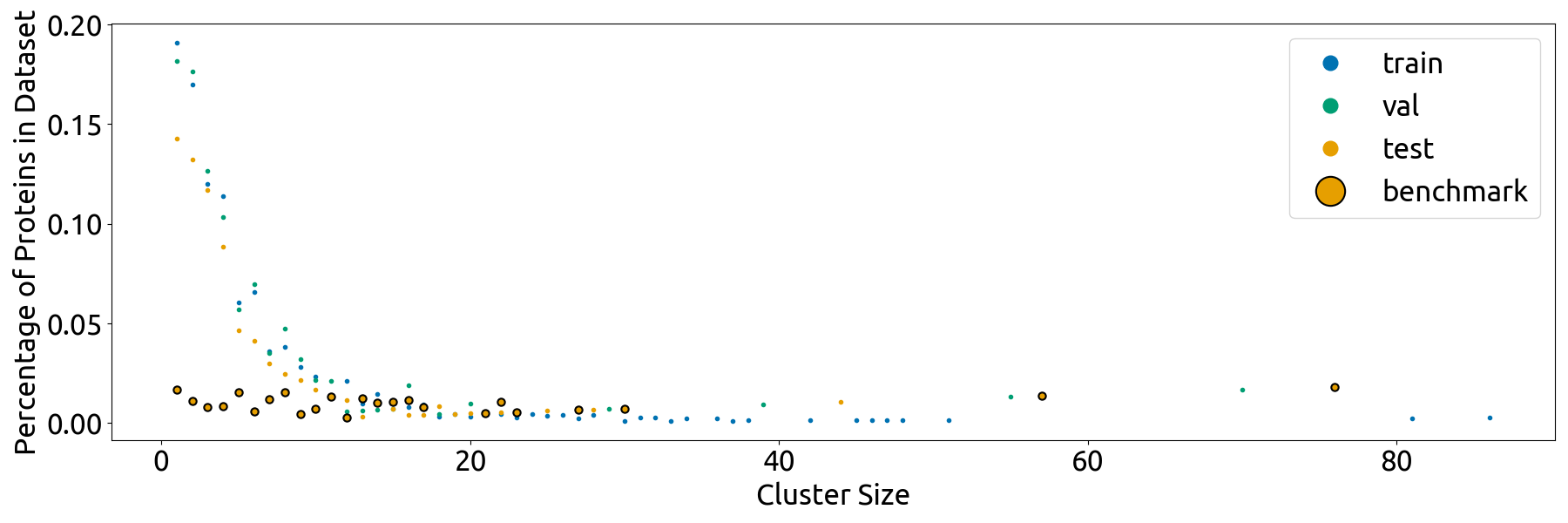
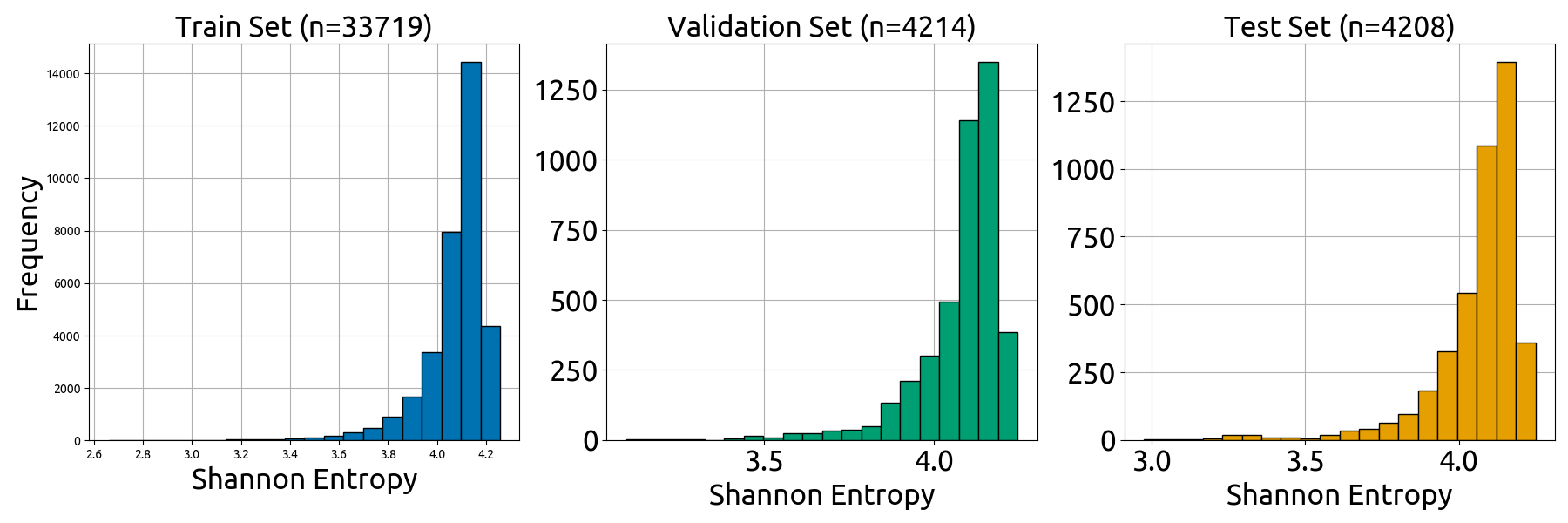
- the **`split_vis`** folder, which contains all visualizations in Fig. S4 and the data that was directly plotted in these visualizations (`*_source_data.csv` files). Note that the individual subplots have slightly different dimensions than they do in the combined Fig. S4
|
| 89 |
+
- **`splits/split_vis/combined_plot.png`**: plot displaying the composition of the train, validation, and test splits (Fig. S4).
|
| 90 |
+
- **`splits/split_vis/length_distributions.png`**: plot displaying the length distributions of the train, validation, and test splits (Fig. 4A)
|
| 91 |
+
- **`splits/split_vis/shannon_entropy_plot.png`**: plot displaying the Shannon entropy distributions of train, validation, and test sets (Fig. 4B)
|
| 92 |
+
- **`splits/split_vis/scatterplot.png`**: plot displaying the cluster size distributions of the train, validation, and test sets (Fig. 4C)
|
| 93 |
+
- **`splits/split_vis/aa_comp.png`**: plot displaying the amino acid composition of the train, validation, and test splits (Fig. S4D).
|
| 94 |
|
| 95 |
### BLAST
|
| 96 |
We ran BLAST to get the best alignment of each sequence in FusOn-DB to a protein in SwissProt. See the README in the `blast` folder for more details.
|
fuson_plm/data/blast/README.md
CHANGED
|
@@ -30,6 +30,7 @@ data/
|
|
| 30 |
├── best_htg_alignments_swissprot_seqs.pkl
|
| 31 |
├── ht_uniprot_query.txt
|
| 32 |
└── figures/
|
|
|
|
| 33 |
├── identities_hist.png
|
| 34 |
├── blast_fusions.py
|
| 35 |
├── extract_blast_seqs.py
|
|
@@ -40,7 +41,7 @@ data/
|
|
| 40 |
|
| 41 |
- **`blast_fusions.py`**: script that will prepare FusOn-DB for BLAST, run BLAST against SwissProt (given you've installed BLAST software properly), extract top alignments and calculate statistics on the BLAST results, and make results plots. Print statements in this script create the log file `fusion_blast_log.txt`.
|
| 42 |
- **`extract_blast_seqs.py`**: script that will extract sequences of all the head/tail proteins that formed the best alignment during BLAST, directly from the SwissProt BLAST database. Creates the file `blast_outputs/best_htg_alignments_swissprot_seqs.pkl`.
|
| 43 |
-
- **`plot.py`**: script to make the plot found at `figures/identities_hist.png`. This plot displays the maximum % identity of each fusion oncoprotein sequence with a SwissProt sequence, based on BLAST. This plot is also automatically created by `blast_fusions.py`.
|
| 44 |
- **`fuson_ht_db.csv`**: Database that merges FusOn-DB (`/*/FusOn-pLM/fuson_plm/data/fuson_db.csv`) with `/*/FusOn-pLM/fuson_plm/data/head_tail_data/htgenes_uniprotids.csv`, which simplifies the process of analyzing BLAST results. In FusOn-DB, certain amino acid sequences are associated with multiple fusion oncoproteins, whose names are comma-separated in the `fusiongenes` column. In `fuson_ht_db.csv`, the `fusiongenes` column is exploded such that exach row only has one fusion gene. Therefore, this database has more rows than FusOn-DB, and some duplicate sequences.
|
| 45 |
|
| 46 |
To run BLAST search and analysis, we recommend using nohup as the process will take a long time.
|
|
|
|
| 30 |
├── best_htg_alignments_swissprot_seqs.pkl
|
| 31 |
├── ht_uniprot_query.txt
|
| 32 |
└── figures/
|
| 33 |
+
├── identities_hist_source_data.png
|
| 34 |
├── identities_hist.png
|
| 35 |
├── blast_fusions.py
|
| 36 |
├── extract_blast_seqs.py
|
|
|
|
| 41 |
|
| 42 |
- **`blast_fusions.py`**: script that will prepare FusOn-DB for BLAST, run BLAST against SwissProt (given you've installed BLAST software properly), extract top alignments and calculate statistics on the BLAST results, and make results plots. Print statements in this script create the log file `fusion_blast_log.txt`.
|
| 43 |
- **`extract_blast_seqs.py`**: script that will extract sequences of all the head/tail proteins that formed the best alignment during BLAST, directly from the SwissProt BLAST database. Creates the file `blast_outputs/best_htg_alignments_swissprot_seqs.pkl`.
|
| 44 |
+
- **`plot.py`**: script to make the plot found at `figures/identities_hist.png` (Fig. 1B histogram). The exact data plotted in this histogram is in `figures/identities_hist_source_data`. This plot displays the maximum % identity of each fusion oncoprotein sequence with a SwissProt sequence, based on BLAST. This plot is also automatically created by `blast_fusions.py`.
|
| 45 |
- **`fuson_ht_db.csv`**: Database that merges FusOn-DB (`/*/FusOn-pLM/fuson_plm/data/fuson_db.csv`) with `/*/FusOn-pLM/fuson_plm/data/head_tail_data/htgenes_uniprotids.csv`, which simplifies the process of analyzing BLAST results. In FusOn-DB, certain amino acid sequences are associated with multiple fusion oncoproteins, whose names are comma-separated in the `fusiongenes` column. In `fuson_ht_db.csv`, the `fusiongenes` column is exploded such that exach row only has one fusion gene. Therefore, this database has more rows than FusOn-DB, and some duplicate sequences.
|
| 46 |
|
| 47 |
To run BLAST search and analysis, we recommend using nohup as the process will take a long time.
|
model/model.pth → fuson_plm/data/blast/figures/identities_hist_source_data.csv
RENAMED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5f2219c7ab63205bcc8d3f21a7d1718aa6054179506411bb641e073b739ca2c7
|
| 3 |
+
size 691452
|
fuson_plm/data/blast/plot.py
CHANGED
|
@@ -19,10 +19,16 @@ def plot_pos_or_id_pcnt_hist(data, column_name, save_path=None, ax=None):
|
|
| 19 |
fig, ax = plt.subplots(figsize=(10, 7))
|
| 20 |
|
| 21 |
# Make the sample data
|
| 22 |
-
data = data[['aa_seq_len', column_name]].dropna() # only keep those with alignments
|
| 23 |
data[column_name] = data[column_name]*100 # so it's %
|
| 24 |
data[f"{column_name} Percent Coverage"] = data[column_name] / data['aa_seq_len']
|
| 25 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 26 |
# Calculate the mean and median of the percent coverage
|
| 27 |
mean_coverage = data[f"{column_name} Percent Coverage"].mean()
|
| 28 |
median_coverage = data[f"{column_name} Percent Coverage"].median()
|
|
|
|
| 19 |
fig, ax = plt.subplots(figsize=(10, 7))
|
| 20 |
|
| 21 |
# Make the sample data
|
| 22 |
+
data = data[['seq_id','aa_seq_len', column_name]].dropna() # only keep those with alignments
|
| 23 |
data[column_name] = data[column_name]*100 # so it's %
|
| 24 |
data[f"{column_name} Percent Coverage"] = data[column_name] / data['aa_seq_len']
|
| 25 |
|
| 26 |
+
# Save this sample data
|
| 27 |
+
source_data_save_path = save_path.replace(".png","_source_data.csv")
|
| 28 |
+
source_data = data[['seq_id',f"{column_name} Percent Coverage"]].sort_values(by=f"{column_name} Percent Coverage",ascending=True)
|
| 29 |
+
source_data[f"{column_name} Percent Coverage"] = source_data[f"{column_name} Percent Coverage"].round(3)
|
| 30 |
+
source_data.to_csv(source_data_save_path,index=False)
|
| 31 |
+
|
| 32 |
# Calculate the mean and median of the percent coverage
|
| 33 |
mean_coverage = data[f"{column_name} Percent Coverage"].mean()
|
| 34 |
median_coverage = data[f"{column_name} Percent Coverage"].median()
|
fuson_plm/data/split_vis.py
CHANGED
|
@@ -6,312 +6,8 @@ import pickle
|
|
| 6 |
import pandas as pd
|
| 7 |
import os
|
| 8 |
from fuson_plm.utils.logging import log_update
|
| 9 |
-
from fuson_plm.utils.visualizing import set_font
|
| 10 |
|
| 11 |
-
def calculate_aa_composition(sequences):
|
| 12 |
-
composition = {}
|
| 13 |
-
total_length = sum([len(seq) for seq in sequences])
|
| 14 |
-
|
| 15 |
-
for seq in sequences:
|
| 16 |
-
for aa in seq:
|
| 17 |
-
if aa in composition:
|
| 18 |
-
composition[aa] += 1
|
| 19 |
-
else:
|
| 20 |
-
composition[aa] = 1
|
| 21 |
-
|
| 22 |
-
# Convert counts to relative frequency
|
| 23 |
-
for aa in composition:
|
| 24 |
-
composition[aa] /= total_length
|
| 25 |
-
|
| 26 |
-
return composition
|
| 27 |
-
|
| 28 |
-
def calculate_shannon_entropy(sequence):
|
| 29 |
-
"""
|
| 30 |
-
Calculate the Shannon entropy for a given sequence.
|
| 31 |
-
|
| 32 |
-
Args:
|
| 33 |
-
sequence (str): A sequence of characters (e.g., amino acids or nucleotides).
|
| 34 |
-
|
| 35 |
-
Returns:
|
| 36 |
-
float: Shannon entropy value.
|
| 37 |
-
"""
|
| 38 |
-
bases = set(sequence)
|
| 39 |
-
counts = [sequence.count(base) for base in bases]
|
| 40 |
-
return entropy(counts, base=2)
|
| 41 |
-
|
| 42 |
-
def visualize_splits_hist(train_lengths, val_lengths, test_lengths, colormap, savepath=f'../data/splits/length_distributions.png', axes=None):
|
| 43 |
-
log_update('\nMaking histogram of length distributions')
|
| 44 |
-
# Create a figure and axes with 1 row and 3 columns
|
| 45 |
-
if axes is None:
|
| 46 |
-
fig, axes = plt.subplots(1, 3, figsize=(18, 6))
|
| 47 |
-
|
| 48 |
-
# Unpack the labels and titles
|
| 49 |
-
xlabel, ylabel = ['Sequence Length (AA)', 'Frequency']
|
| 50 |
-
|
| 51 |
-
# Plot the first histogram
|
| 52 |
-
axes[0].hist(train_lengths, bins=20, edgecolor='k',color=colormap['train'])
|
| 53 |
-
axes[0].set_xlabel(xlabel, fontsize=24)
|
| 54 |
-
axes[0].set_ylabel(ylabel, fontsize=24)
|
| 55 |
-
axes[0].set_title(f'Train Set Length Distribution (n={len(train_lengths)})', fontsize=24)
|
| 56 |
-
axes[0].grid(True)
|
| 57 |
-
axes[0].set_axisbelow(True)
|
| 58 |
-
axes[0].tick_params(axis='x', labelsize=24) # Customize x-axis tick label size
|
| 59 |
-
axes[0].tick_params(axis='y', labelsize=24) # Customize y-axis tick label size
|
| 60 |
-
|
| 61 |
-
|
| 62 |
-
# Plot the second histogram
|
| 63 |
-
axes[1].hist(val_lengths, bins=20, edgecolor='k',color=colormap['val'])
|
| 64 |
-
axes[1].set_xlabel(xlabel, fontsize=24)
|
| 65 |
-
axes[1].set_ylabel(ylabel, fontsize=24)
|
| 66 |
-
axes[1].set_title(f'Validation Set Length Distribution (n={len(val_lengths)})', fontsize=24)
|
| 67 |
-
axes[1].grid(True)
|
| 68 |
-
axes[1].set_axisbelow(True)
|
| 69 |
-
axes[1].tick_params(axis='x', labelsize=24)
|
| 70 |
-
axes[1].tick_params(axis='y', labelsize=24)
|
| 71 |
-
|
| 72 |
-
# Plot the third histogram
|
| 73 |
-
axes[2].hist(test_lengths, bins=20, edgecolor='k',color=colormap['test'])
|
| 74 |
-
axes[2].set_xlabel(xlabel, fontsize=24)
|
| 75 |
-
axes[2].set_ylabel(ylabel, fontsize=24)
|
| 76 |
-
axes[2].set_title(f'Test Set Length Distribution (n={len(test_lengths)})', fontsize=24)
|
| 77 |
-
axes[2].grid(True)
|
| 78 |
-
axes[2].set_axisbelow(True)
|
| 79 |
-
axes[2].tick_params(axis='x', labelsize=24)
|
| 80 |
-
axes[2].tick_params(axis='y', labelsize=24)
|
| 81 |
-
|
| 82 |
-
# Adjust layout
|
| 83 |
-
if savepath is not None:
|
| 84 |
-
plt.tight_layout()
|
| 85 |
-
|
| 86 |
-
# Save the figure
|
| 87 |
-
plt.savefig(savepath)
|
| 88 |
-
|
| 89 |
-
def visualize_splits_scatter(train_clusters, val_clusters, test_clusters, benchmark_cluster_reps, colormap, savepath='../data/splits/scatterplot.png', ax=None):
|
| 90 |
-
log_update("\nMaking scatterplot with distribution of cluster sizes across train, test, and val")
|
| 91 |
-
# Make grouped versions of these DataFrames for size analysis
|
| 92 |
-
train_clustersgb = train_clusters.groupby('representative seq_id')['member seq_id'].count().reset_index().rename(columns={'member seq_id':'member count'})
|
| 93 |
-
val_clustersgb = val_clusters.groupby('representative seq_id')['member seq_id'].count().reset_index().rename(columns={'member seq_id':'member count'})
|
| 94 |
-
test_clustersgb = test_clusters.groupby('representative seq_id')['member seq_id'].count().reset_index().rename(columns={'member seq_id':'member count'})
|
| 95 |
-
|
| 96 |
-
# Isolate benchmark-containing clusters so their contribution can be plotted separately
|
| 97 |
-
total_test_proteins = sum(test_clustersgb['member count'])
|
| 98 |
-
test_clustersgb['benchmark cluster'] = test_clustersgb['representative seq_id'].isin(benchmark_cluster_reps)
|
| 99 |
-
benchmark_clustersgb = test_clustersgb.loc[test_clustersgb['benchmark cluster']].reset_index(drop=True)
|
| 100 |
-
test_clustersgb = test_clustersgb.loc[test_clustersgb['benchmark cluster']==False].reset_index(drop=True)
|
| 101 |
-
|
| 102 |
-
# Convert them to value counts
|
| 103 |
-
train_clustersgb = train_clustersgb['member count'].value_counts().reset_index().rename(columns={'index':'cluster size (n_members)','member count': 'n_clusters'})
|
| 104 |
-
val_clustersgb = val_clustersgb['member count'].value_counts().reset_index().rename(columns={'index':'cluster size (n_members)','member count': 'n_clusters'})
|
| 105 |
-
test_clustersgb = test_clustersgb['member count'].value_counts().reset_index().rename(columns={'index':'cluster size (n_members)','member count': 'n_clusters'})
|
| 106 |
-
benchmark_clustersgb = benchmark_clustersgb['member count'].value_counts().reset_index().rename(columns={'index':'cluster size (n_members)','member count': 'n_clusters'})
|
| 107 |
-
|
| 108 |
-
# Get the percentage of each dataset that's made of each cluster size
|
| 109 |
-
train_clustersgb['n_proteins'] = train_clustersgb['cluster size (n_members)']*train_clustersgb['n_clusters'] # proteins per cluster * n clusters = # proteins
|
| 110 |
-
train_clustersgb['percent_proteins'] = train_clustersgb['n_proteins']/sum(train_clustersgb['n_proteins'])
|
| 111 |
-
val_clustersgb['n_proteins'] = val_clustersgb['cluster size (n_members)']*val_clustersgb['n_clusters']
|
| 112 |
-
val_clustersgb['percent_proteins'] = val_clustersgb['n_proteins']/sum(val_clustersgb['n_proteins'])
|
| 113 |
-
test_clustersgb['n_proteins'] = test_clustersgb['cluster size (n_members)']*test_clustersgb['n_clusters']
|
| 114 |
-
test_clustersgb['percent_proteins'] = test_clustersgb['n_proteins']/total_test_proteins
|
| 115 |
-
benchmark_clustersgb['n_proteins'] = benchmark_clustersgb['cluster size (n_members)']*benchmark_clustersgb['n_clusters']
|
| 116 |
-
benchmark_clustersgb['percent_proteins'] = benchmark_clustersgb['n_proteins']/total_test_proteins
|
| 117 |
-
|
| 118 |
-
# Specially mark the benchmark clusters because these can't be reallocated
|
| 119 |
-
if ax is None:
|
| 120 |
-
fig, ax = plt.subplots(figsize=(18, 6))
|
| 121 |
-
|
| 122 |
-
ax.plot(train_clustersgb['cluster size (n_members)'],train_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['train'],label='train')
|
| 123 |
-
ax.plot(val_clustersgb['cluster size (n_members)'],val_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['val'],label='val')
|
| 124 |
-
ax.plot(test_clustersgb['cluster size (n_members)'],test_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['test'],label='test')
|
| 125 |
-
ax.plot(benchmark_clustersgb['cluster size (n_members)'],benchmark_clustersgb['percent_proteins'],
|
| 126 |
-
marker='o',
|
| 127 |
-
linestyle='None',
|
| 128 |
-
markerfacecolor=colormap['test'], # fill same as test
|
| 129 |
-
markeredgecolor='black', # outline black
|
| 130 |
-
markeredgewidth=1.5,
|
| 131 |
-
label='benchmark'
|
| 132 |
-
)
|
| 133 |
-
ax.set_ylabel('Percentage of Proteins in Dataset', fontsize=24)
|
| 134 |
-
ax.set_xlabel('Cluster Size', fontsize=24)
|
| 135 |
-
ax.tick_params(axis='x', labelsize=24) # Customize x-axis tick label size
|
| 136 |
-
ax.tick_params(axis='y', labelsize=24) # Customize y-axis tick label size
|
| 137 |
-
|
| 138 |
-
ax.legend(fontsize=24,markerscale=4)
|
| 139 |
-
|
| 140 |
-
# save the figure
|
| 141 |
-
if savepath is not None:
|
| 142 |
-
plt.tight_layout()
|
| 143 |
-
plt.savefig(savepath)
|
| 144 |
-
log_update(f"\tSaved figure to {savepath}")
|
| 145 |
-
|
| 146 |
-
def get_avg_embeddings_for_tsne(train_sequences, val_sequences, test_sequences, embedding_path='fuson_db_embeddings/fuson_db_esm2_t33_650M_UR50D_avg_embeddings.pkl'):
|
| 147 |
-
embeddings = {}
|
| 148 |
-
|
| 149 |
-
try:
|
| 150 |
-
with open(embedding_path, 'rb') as f:
|
| 151 |
-
embeddings = pickle.load(f)
|
| 152 |
-
|
| 153 |
-
train_embeddings = [v for k, v in embeddings.items() if k in train_sequences]
|
| 154 |
-
val_embeddings = [v for k, v in embeddings.items() if k in val_sequences]
|
| 155 |
-
test_embeddings = [v for k, v in embeddings.items() if k in test_sequences]
|
| 156 |
-
|
| 157 |
-
return train_embeddings, val_embeddings, test_embeddings
|
| 158 |
-
except:
|
| 159 |
-
print("could not open embeddings")
|
| 160 |
-
|
| 161 |
-
|
| 162 |
-
def visualize_splits_tsne(train_sequences, val_sequences, test_sequences, colormap, esm_type="esm2_t33_650M_UR50D", embedding_path="fuson_db_embeddings/fuson_db_esm2_t33_650M_UR50D_avg_embeddings.pkl", savepath='../data/splits/tsne_plot.png',ax=None):
|
| 163 |
-
"""
|
| 164 |
-
Generate a t-SNE plot of embeddings for train, test, and validation.
|
| 165 |
-
"""
|
| 166 |
-
log_update('\nMaking t-SNE plot of train, val, and test embeddings')
|
| 167 |
-
# Combine the embeddings into one array
|
| 168 |
-
train_embeddings, val_embeddings, test_embeddings = get_avg_embeddings_for_tsne(train_sequences, val_sequences, test_sequences, embedding_path=embedding_path)
|
| 169 |
-
embeddings = np.concatenate([train_embeddings, val_embeddings, test_embeddings])
|
| 170 |
-
|
| 171 |
-
# Labels for the embeddings
|
| 172 |
-
labels = ['train'] * len(train_embeddings) + ['val'] * len(val_embeddings) + ['test'] * len(test_embeddings)
|
| 173 |
-
|
| 174 |
-
# Perform t-SNE
|
| 175 |
-
tsne = TSNE(n_components=2, random_state=42)
|
| 176 |
-
tsne_results = tsne.fit_transform(embeddings)
|
| 177 |
-
|
| 178 |
-
# Convert t-SNE results into a DataFrame
|
| 179 |
-
tsne_df = pd.DataFrame(data=tsne_results, columns=['TSNE_1', 'TSNE_2'])
|
| 180 |
-
tsne_df['label'] = labels
|
| 181 |
-
|
| 182 |
-
# Plotting
|
| 183 |
-
if ax is None:
|
| 184 |
-
fig, ax = plt.subplots(figsize=(10, 8))
|
| 185 |
-
|
| 186 |
-
# Scatter plot for each set
|
| 187 |
-
for label, color in colormap.items():
|
| 188 |
-
subset = tsne_df[tsne_df['label'] == label].reset_index(drop=True)
|
| 189 |
-
ax.scatter(subset['TSNE_1'], subset['TSNE_2'], c=color, label=label.capitalize(), alpha=0.6)
|
| 190 |
-
|
| 191 |
-
ax.set_title(f't-SNE of {esm_type} Embeddings')
|
| 192 |
-
ax.set_xlabel('t-SNE Dimension 1')
|
| 193 |
-
ax.set_ylabel('t-SNE Dimension 2')
|
| 194 |
-
ax.legend(fontsize=24, markerscale=2)
|
| 195 |
-
ax.grid(True)
|
| 196 |
-
|
| 197 |
-
# Save the figure if savepath is provided
|
| 198 |
-
if savepath:
|
| 199 |
-
plt.tight_layout()
|
| 200 |
-
fig.savefig(savepath)
|
| 201 |
-
|
| 202 |
-
def visualize_splits_shannon_entropy(train_sequences, val_sequences, test_sequences, colormap, savepath='../data/splits/shannon_entropy_plot.png',axes=None):
|
| 203 |
-
"""
|
| 204 |
-
Generate Shannon entropy plots for train, validation, and test sets.
|
| 205 |
-
"""
|
| 206 |
-
log_update('\nMaking histogram of Shannon Entropy distributions')
|
| 207 |
-
train_entropy = [calculate_shannon_entropy(seq) for seq in train_sequences]
|
| 208 |
-
val_entropy = [calculate_shannon_entropy(seq) for seq in val_sequences]
|
| 209 |
-
test_entropy = [calculate_shannon_entropy(seq) for seq in test_sequences]
|
| 210 |
-
|
| 211 |
-
if axes is None:
|
| 212 |
-
fig, axes = plt.subplots(1, 3, figsize=(18, 6))
|
| 213 |
-
|
| 214 |
-
axes[0].hist(train_entropy, bins=20, edgecolor='k', color=colormap['train'])
|
| 215 |
-
axes[0].set_title(f'Train Set (n={len(train_entropy)})', fontsize=24)
|
| 216 |
-
axes[0].set_xlabel('Shannon Entropy', fontsize=24)
|
| 217 |
-
axes[0].set_ylabel('Frequency', fontsize=24)
|
| 218 |
-
axes[0].grid(True)
|
| 219 |
-
axes[0].set_axisbelow(True)
|
| 220 |
-
axes[0].tick_params(axis='x', labelsize=24)
|
| 221 |
-
axes[0].tick_params(axis='y', labelsize=24)
|
| 222 |
-
|
| 223 |
-
axes[1].hist(val_entropy, bins=20, edgecolor='k', color=colormap['val'])
|
| 224 |
-
axes[1].set_title(f'Validation Set (n={len(val_entropy)})', fontsize=24)
|
| 225 |
-
axes[1].set_xlabel('Shannon Entropy', fontsize=24)
|
| 226 |
-
axes[1].grid(True)
|
| 227 |
-
axes[1].set_axisbelow(True)
|
| 228 |
-
axes[1].tick_params(axis='x', labelsize=24)
|
| 229 |
-
axes[1].tick_params(axis='y', labelsize=24)
|
| 230 |
-
|
| 231 |
-
axes[2].hist(test_entropy, bins=20, edgecolor='k', color=colormap['test'])
|
| 232 |
-
axes[2].set_title(f'Test Set (n={len(test_entropy)})', fontsize=24)
|
| 233 |
-
axes[2].set_xlabel('Shannon Entropy', fontsize=24)
|
| 234 |
-
axes[2].grid(True)
|
| 235 |
-
axes[2].set_axisbelow(True)
|
| 236 |
-
axes[2].tick_params(axis='x', labelsize=24)
|
| 237 |
-
axes[2].tick_params(axis='y', labelsize=24)
|
| 238 |
-
|
| 239 |
-
if savepath is not None:
|
| 240 |
-
plt.tight_layout()
|
| 241 |
-
plt.savefig(savepath)
|
| 242 |
-
|
| 243 |
-
def visualize_splits_aa_composition(train_sequences, val_sequences, test_sequences,colormap, savepath='../data/splits/aa_comp.png',ax=None):
|
| 244 |
-
log_update('\nMaking bar plot of AA composition across each set')
|
| 245 |
-
train_comp = calculate_aa_composition(train_sequences)
|
| 246 |
-
val_comp = calculate_aa_composition(val_sequences)
|
| 247 |
-
test_comp = calculate_aa_composition(test_sequences)
|
| 248 |
-
|
| 249 |
-
# Create DataFrame
|
| 250 |
-
comp_df = pd.DataFrame([train_comp, val_comp, test_comp], index=['train', 'val', 'test']).T
|
| 251 |
-
colors = [colormap[col] for col in comp_df.columns]
|
| 252 |
-
|
| 253 |
-
# Plotting
|
| 254 |
-
#fig, ax = plt.subplots(figsize=(12, 6))
|
| 255 |
-
if ax is None:
|
| 256 |
-
fig, ax = plt.subplots(figsize=(12, 6))
|
| 257 |
-
else:
|
| 258 |
-
fig = ax.get_figure()
|
| 259 |
-
|
| 260 |
-
comp_df.plot(kind='bar', color=colors, ax=ax)
|
| 261 |
-
ax.set_title('Amino Acid Composition Across Datasets', fontsize=24)
|
| 262 |
-
ax.set_xlabel('Amino Acid', fontsize=24)
|
| 263 |
-
ax.set_ylabel('Relative Frequency', fontsize=24)
|
| 264 |
-
ax.tick_params(axis='x', labelsize=24) # Customize x-axis tick label size
|
| 265 |
-
ax.tick_params(axis='y', labelsize=24) # Customize y-axis tick label size
|
| 266 |
-
ax.legend(fontsize=16, markerscale=2)
|
| 267 |
-
|
| 268 |
-
if savepath is not None:
|
| 269 |
-
fig.savefig(savepath)
|
| 270 |
-
|
| 271 |
-
def visualize_splits(train_clusters, val_clusters, test_clusters, benchmark_cluster_reps, train_color='#0072B2',val_color='#009E73',test_color='#E69F00',esm_embeddings_path=None, onehot_embeddings_path=None):
|
| 272 |
-
colormap = {
|
| 273 |
-
'train': train_color,
|
| 274 |
-
'val': val_color,
|
| 275 |
-
'test': test_color
|
| 276 |
-
}
|
| 277 |
-
# Add columns for plotting
|
| 278 |
-
train_clusters['member length'] = train_clusters['member seq'].str.len()
|
| 279 |
-
val_clusters['member length'] = val_clusters['member seq'].str.len()
|
| 280 |
-
test_clusters['member length'] = test_clusters['member seq'].str.len()
|
| 281 |
-
|
| 282 |
-
# Prepare lengths and seqs for plotting
|
| 283 |
-
train_lengths = train_clusters['member length'].tolist()
|
| 284 |
-
val_lengths = val_clusters['member length'].tolist()
|
| 285 |
-
test_lengths = test_clusters['member length'].tolist()
|
| 286 |
-
train_sequences = train_clusters['member seq'].tolist()
|
| 287 |
-
val_sequences = val_clusters['member seq'].tolist()
|
| 288 |
-
test_sequences = test_clusters['member seq'].tolist()
|
| 289 |
-
|
| 290 |
-
# Create a combined figure with 3 rows and 3 columns
|
| 291 |
-
fig_combined, axs = plt.subplots(3, 3, figsize=(24, 18))
|
| 292 |
-
|
| 293 |
-
# Make the three visualization plots for saving TOGETHER
|
| 294 |
-
visualize_splits_hist(train_lengths,val_lengths,test_lengths,colormap, savepath=None,axes=axs[0])
|
| 295 |
-
visualize_splits_shannon_entropy(train_sequences,val_sequences,test_sequences,colormap,savepath=None,axes=axs[1])
|
| 296 |
-
visualize_splits_scatter(train_clusters, val_clusters, test_clusters, benchmark_cluster_reps, colormap, savepath=None, ax=axs[2, 0])
|
| 297 |
-
visualize_splits_aa_composition(train_sequences,val_sequences,test_sequences, colormap, savepath=None, ax=axs[2, 1])
|
| 298 |
-
if not(esm_embeddings_path is None) and os.path.exists(esm_embeddings_path):
|
| 299 |
-
visualize_splits_tsne(train_sequences, val_sequences, test_sequences, colormap, savepath=None, ax=axs[2, 2])
|
| 300 |
-
else:
|
| 301 |
-
# Leave the last subplot blank
|
| 302 |
-
axs[2, 2].axis('off')
|
| 303 |
-
|
| 304 |
-
plt.tight_layout()
|
| 305 |
-
fig_combined.savefig('../data/splits/combined_plot.png')
|
| 306 |
-
|
| 307 |
-
# Make the three visualization plots for saving separately
|
| 308 |
-
visualize_splits_hist(train_clusters['member length'].tolist(), val_clusters['member length'].tolist(), test_clusters['member length'].tolist(),colormap)
|
| 309 |
-
visualize_splits_scatter(train_clusters, val_clusters, test_clusters, benchmark_cluster_reps, colormap)
|
| 310 |
-
visualize_splits_aa_composition(train_clusters['member seq'].tolist(), val_clusters['member seq'].tolist(), test_clusters['member seq'].tolist(),colormap)
|
| 311 |
-
visualize_splits_shannon_entropy(train_sequences,val_sequences,test_sequences,colormap)
|
| 312 |
-
if not(esm_embeddings_path is None) and os.path.exists(esm_embeddings_path):
|
| 313 |
-
visualize_splits_tsne(train_clusters['member seq'].tolist(), val_clusters['member seq'].tolist(), test_clusters['member seq'].tolist(),colormap)
|
| 314 |
-
|
| 315 |
def main():
|
| 316 |
set_font()
|
| 317 |
train_clusters = pd.read_csv('splits/train_cluster_split.csv')
|
|
@@ -326,8 +22,19 @@ def main():
|
|
| 326 |
# Use benchmark_seq_ids to find which clusters contain benchmark sequences.
|
| 327 |
benchmark_cluster_reps = clusters.loc[clusters['member seq_id'].isin(benchmark_seq_ids)]['representative seq_id'].unique().tolist()
|
| 328 |
|
| 329 |
-
visualize_splits(train_clusters, val_clusters, test_clusters, benchmark_cluster_reps
|
| 330 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 331 |
|
| 332 |
if __name__ == "__main__":
|
| 333 |
main()
|
|
|
|
| 6 |
import pandas as pd
|
| 7 |
import os
|
| 8 |
from fuson_plm.utils.logging import log_update
|
| 9 |
+
from fuson_plm.utils.visualizing import set_font, visualize_splits
|
| 10 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 11 |
def main():
|
| 12 |
set_font()
|
| 13 |
train_clusters = pd.read_csv('splits/train_cluster_split.csv')
|
|
|
|
| 22 |
# Use benchmark_seq_ids to find which clusters contain benchmark sequences.
|
| 23 |
benchmark_cluster_reps = clusters.loc[clusters['member seq_id'].isin(benchmark_seq_ids)]['representative seq_id'].unique().tolist()
|
| 24 |
|
| 25 |
+
visualize_splits(train_clusters, val_clusters, test_clusters, benchmark_cluster_reps)
|
| 26 |
+
|
| 27 |
+
## Add seq_id to every source data file that is saved from visualize_splits
|
| 28 |
+
seq_to_id_dict = dict(zip(fuson_db['aa_seq'],fuson_db['seq_id']))
|
| 29 |
+
files_to_edit = os.listdir("splits/split_vis")
|
| 30 |
+
files_to_edit = [x for x in files_to_edit if x[-4::]==".csv"]
|
| 31 |
+
log_update(f"Adding seq_ids to the following files: {files_to_edit}")
|
| 32 |
+
|
| 33 |
+
for fname in files_to_edit:
|
| 34 |
+
source_data_file = pd.read_csv(f"splits/split_vis/{fname}")
|
| 35 |
+
if "sequence" in list(source_data_file.columns):
|
| 36 |
+
source_data_file["seq_id"] = source_data_file["sequence"].map(seq_to_id_dict)
|
| 37 |
+
source_data_file.drop(columns=['sequence']).to_csv(f"splits/split_vis/{fname}",index=False)
|
| 38 |
|
| 39 |
if __name__ == "__main__":
|
| 40 |
main()
|
fuson_plm/data/splits/combined_plot.png
DELETED
|
Binary file (267 kB)
|
|
|
fuson_plm/data/splits/split_vis/aa_comp.png
ADDED
|
fuson_plm/data/splits/split_vis/aa_comp_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:c2f71076771c047787076795f088ec99c532fe13a901890c29355431d5cbf428
|
| 3 |
+
size 1273
|
fuson_plm/data/splits/split_vis/combined_plot.png
ADDED
|
fuson_plm/data/splits/split_vis/length_distributions.png
ADDED

|
fuson_plm/data/splits/split_vis/scatterplot.png
ADDED
|
fuson_plm/data/splits/split_vis/scatterplot_benchmark_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:45b3b580146c214c76fec277ac721b2de0f1f9a5f0c8096dad13f39340d15da1
|
| 3 |
+
size 755
|
fuson_plm/data/splits/split_vis/scatterplot_test_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:1e888045ff3c6824e98ddb3c25f45cdae80d06bfddf7878c1309ef7d47da9ce8
|
| 3 |
+
size 794
|
fuson_plm/data/splits/split_vis/scatterplot_train_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8047af5c61b9238654dbd4d3f693fad68e09ece90fb72fb5879530bd17969d3d
|
| 3 |
+
size 1396
|
fuson_plm/data/splits/split_vis/scatterplot_val_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5802d962e638004536d5473ccb7d5f3150a481d1295e1abf9fed55fe28311aea
|
| 3 |
+
size 819
|
fuson_plm/data/splits/split_vis/shannon_entropy_plot.png
ADDED
|
fuson_plm/data/splits/split_vis/shannon_entropy_plot_test_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:51bb7926f16e082974f2693b67b77d7d7246e63766cb76ecf9f0638fe9656670
|
| 3 |
+
size 112646
|
fuson_plm/data/splits/split_vis/shannon_entropy_plot_train_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4dc397807e7f6114d8cdd571efb779ac9d98f76f1e5f0c9a872fe1b355453d54
|
| 3 |
+
size 902131
|
fuson_plm/data/splits/split_vis/shannon_entropy_plot_val_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:d25a06e2ccf701d7299922a2978ea478dd0fd78339e852060628a71bbbe024a9
|
| 3 |
+
size 112779
|
fuson_plm/data/splits/split_vis/test_lengths_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:2ac21f806a235f769afe107ed8bc89476d3f9af66c30ff2b1c6ff55da9a8a1f1
|
| 3 |
+
size 54367
|
fuson_plm/data/splits/split_vis/train_lengths_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:146c64a03c446cc7c5a59b4d72edd099b526e1f4c9e777a1cbed7f6dd410a3b6
|
| 3 |
+
size 435911
|
fuson_plm/data/splits/split_vis/val_lengths_source_data.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5fd936ad5daed767e3815dac6315d92abcabb4fe506c169e6a958031a4fd2d97
|
| 3 |
+
size 54478
|
fuson_plm/utils/visualizing.py
CHANGED
|
@@ -34,11 +34,12 @@ def set_font():
|
|
| 34 |
# Set the font family globally to Ubuntu
|
| 35 |
plt.rcParams['font.family'] = regular_font.get_name()
|
| 36 |
|
| 37 |
-
# Set the
|
|
|
|
| 38 |
plt.rcParams['mathtext.fontset'] = 'custom'
|
| 39 |
plt.rcParams['mathtext.rm'] = regular_font.get_name()
|
| 40 |
-
plt.rcParams['mathtext.it'] =
|
| 41 |
-
plt.rcParams['mathtext.bf'] =
|
| 42 |
|
| 43 |
global default_color_map
|
| 44 |
default_color_map = {
|
|
@@ -98,7 +99,7 @@ def calculate_shannon_entropy(sequence):
|
|
| 98 |
counts = [sequence.count(base) for base in bases]
|
| 99 |
return entropy(counts, base=2)
|
| 100 |
|
| 101 |
-
def visualize_splits_hist(train_lengths=None, val_lengths=None, test_lengths=None, colormap=None, savepath=f'splits/length_distributions.png', axes=None):
|
| 102 |
"""
|
| 103 |
Works to plot train, val, test; train, val; or train, test
|
| 104 |
"""
|
|
@@ -118,7 +119,7 @@ def visualize_splits_hist(train_lengths=None, val_lengths=None, test_lengths=Non
|
|
| 118 |
total_plots-=1
|
| 119 |
|
| 120 |
# Create a figure and axes with 1 row and 3 columns
|
| 121 |
-
fig_individual, axes_individual = plt.subplots(1, total_plots, figsize=(
|
| 122 |
|
| 123 |
# Set axes list
|
| 124 |
axes_list = [axes_individual] if axes is None else [axes_individual, axes]
|
|
@@ -129,29 +130,35 @@ def visualize_splits_hist(train_lengths=None, val_lengths=None, test_lengths=Non
|
|
| 129 |
for cur_axes in axes_list:
|
| 130 |
# Plot the first histogram
|
| 131 |
cur_axes[0].hist(train_lengths, bins=20, edgecolor='k',color=colormap['train'])
|
| 132 |
-
cur_axes[0].set_xlabel(xlabel)
|
| 133 |
-
cur_axes[0].set_ylabel(ylabel)
|
| 134 |
-
cur_axes[0].set_title(f'Train Set Length Distribution (n={len(train_lengths)})')
|
| 135 |
cur_axes[0].grid(True)
|
| 136 |
cur_axes[0].set_axisbelow(True)
|
|
|
|
|
|
|
| 137 |
|
| 138 |
# Plot the second histogram
|
| 139 |
if not(val_plot_index is None):
|
| 140 |
cur_axes[val_plot_index].hist(val_lengths, bins=20, edgecolor='k',color=colormap['val'])
|
| 141 |
-
cur_axes[val_plot_index].set_xlabel(xlabel)
|
| 142 |
-
cur_axes[val_plot_index].set_ylabel(ylabel)
|
| 143 |
-
cur_axes[val_plot_index].set_title(f'Validation Set Length Distribution (n={len(val_lengths)})')
|
| 144 |
cur_axes[val_plot_index].grid(True)
|
| 145 |
cur_axes[val_plot_index].set_axisbelow(True)
|
|
|
|
|
|
|
| 146 |
|
| 147 |
# Plot the third histogram
|
| 148 |
if not(test_plot_index is None):
|
| 149 |
cur_axes[test_plot_index].hist(test_lengths, bins=20, edgecolor='k',color=colormap['test'])
|
| 150 |
-
cur_axes[test_plot_index].set_xlabel(xlabel)
|
| 151 |
-
cur_axes[test_plot_index].set_ylabel(ylabel)
|
| 152 |
-
cur_axes[test_plot_index].set_title(f'Test Set Length Distribution (n={len(test_lengths)})')
|
| 153 |
cur_axes[test_plot_index].grid(True)
|
| 154 |
cur_axes[test_plot_index].set_axisbelow(True)
|
|
|
|
|
|
|
| 155 |
|
| 156 |
# Adjust layout
|
| 157 |
fig_individual.set_tight_layout(True)
|
|
@@ -160,7 +167,7 @@ def visualize_splits_hist(train_lengths=None, val_lengths=None, test_lengths=Non
|
|
| 160 |
fig_individual.savefig(savepath)
|
| 161 |
log_update(f"\tSaved figure to {savepath}")
|
| 162 |
|
| 163 |
-
def visualize_splits_scatter(train_clusters=None, val_clusters=None, test_clusters=None, benchmark_cluster_reps=None, colormap=None, savepath='splits/scatterplot.png', axes=None):
|
| 164 |
set_font()
|
| 165 |
if colormap is None: colormap=default_color_map
|
| 166 |
|
|
@@ -209,10 +216,13 @@ def visualize_splits_scatter(train_clusters=None, val_clusters=None, test_cluste
|
|
| 209 |
# Specially mark the benchmark clusters because these can't be reallocated
|
| 210 |
for ax in axes_list:
|
| 211 |
ax.plot(train_clustersgb['cluster size (n_members)'],train_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['train'],label='train')
|
|
|
|
| 212 |
if not(val_clusters is None):
|
| 213 |
ax.plot(val_clustersgb['cluster size (n_members)'],val_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['val'],label='val')
|
|
|
|
| 214 |
if not(test_clusters is None):
|
| 215 |
ax.plot(test_clustersgb['cluster size (n_members)'],test_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['test'],label='test')
|
|
|
|
| 216 |
if not(benchmark_cluster_reps is None):
|
| 217 |
ax.plot(benchmark_clustersgb['cluster size (n_members)'],benchmark_clustersgb['percent_proteins'],
|
| 218 |
marker='o',
|
|
@@ -222,8 +232,13 @@ def visualize_splits_scatter(train_clusters=None, val_clusters=None, test_cluste
|
|
| 222 |
markeredgewidth=1.5,
|
| 223 |
label='benchmark'
|
| 224 |
)
|
| 225 |
-
|
| 226 |
-
ax.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 227 |
|
| 228 |
# save the figure
|
| 229 |
fig_individual.set_tight_layout(True)
|
|
@@ -231,7 +246,7 @@ def visualize_splits_scatter(train_clusters=None, val_clusters=None, test_cluste
|
|
| 231 |
log_update(f"\tSaved figure to {savepath}")
|
| 232 |
|
| 233 |
|
| 234 |
-
def visualize_splits_tsne(train_sequences=None, val_sequences=None, test_sequences=None, colormap=None, esm_type="esm2_t33_650M_UR50D", embedding_path="fuson_db_embeddings/fuson_db_esm2_t33_650M_UR50D_avg_embeddings.pkl", savepath='splits/tsne_plot.png',axes=None):
|
| 235 |
set_font()
|
| 236 |
|
| 237 |
if colormap is None: colormap=default_color_map
|
|
@@ -285,7 +300,7 @@ def visualize_splits_tsne(train_sequences=None, val_sequences=None, test_sequenc
|
|
| 285 |
fig_individual.savefig(savepath)
|
| 286 |
log_update(f"\tSaved figure to {savepath}")
|
| 287 |
|
| 288 |
-
def visualize_splits_shannon_entropy(train_sequences=None, val_sequences=None, test_sequences=None, colormap=None, savepath='splits/shannon_entropy_plot.png',axes=None):
|
| 289 |
set_font()
|
| 290 |
"""
|
| 291 |
Generate Shannon entropy plots for train, validation, and test sets.
|
|
@@ -316,31 +331,52 @@ def visualize_splits_shannon_entropy(train_sequences=None, val_sequences=None, t
|
|
| 316 |
|
| 317 |
for ax in axes_list:
|
| 318 |
ax[0].hist(train_entropy, bins=20, edgecolor='k', color=colormap['train'])
|
| 319 |
-
ax[0].set_title(f'Train Set (n={len(train_entropy)})')
|
| 320 |
-
ax[0].set_xlabel('Shannon Entropy')
|
| 321 |
-
ax[0].set_ylabel('Frequency')
|
| 322 |
ax[0].grid(True)
|
| 323 |
ax[0].set_axisbelow(True)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 324 |
|
| 325 |
if not(val_plot_index is None):
|
| 326 |
ax[val_plot_index].hist(val_entropy, bins=20, edgecolor='k', color=colormap['val'])
|
| 327 |
-
ax[val_plot_index].set_title(f'Validation Set (n={len(val_entropy)})')
|
| 328 |
-
ax[val_plot_index].set_xlabel('Shannon Entropy')
|
| 329 |
ax[val_plot_index].grid(True)
|
| 330 |
ax[val_plot_index].set_axisbelow(True)
|
| 331 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 332 |
if not(test_plot_index is None):
|
| 333 |
ax[test_plot_index].hist(test_entropy, bins=20, edgecolor='k', color=colormap['test'])
|
| 334 |
-
ax[test_plot_index].set_title(f'Test Set (n={len(test_entropy)})')
|
| 335 |
-
ax[test_plot_index].set_xlabel('Shannon Entropy')
|
| 336 |
ax[test_plot_index].grid(True)
|
| 337 |
ax[test_plot_index].set_axisbelow(True)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 338 |
|
| 339 |
fig_individual.set_tight_layout(True)
|
| 340 |
fig_individual.savefig(savepath)
|
| 341 |
log_update(f"\tSaved figure to {savepath}")
|
| 342 |
|
| 343 |
-
def visualize_splits_aa_composition(train_sequences=None, val_sequences=None, test_sequences=None, colormap=None, savepath='splits/aa_comp.png',axes=None):
|
| 344 |
set_font()
|
| 345 |
if colormap is None: colormap=default_color_map
|
| 346 |
|
|
@@ -365,13 +401,17 @@ def visualize_splits_aa_composition(train_sequences=None, val_sequences=None, te
|
|
| 365 |
if (val_sequences is None) and not(test_sequences is None):
|
| 366 |
comp_df = pd.DataFrame([train_comp, test_comp], index=['train', 'test']).T
|
| 367 |
colors = [colormap[col] for col in comp_df.columns]
|
|
|
|
| 368 |
|
| 369 |
# Plotting
|
| 370 |
for ax in axes_list:
|
| 371 |
comp_df.plot(kind='bar', color=colors, ax=ax)
|
| 372 |
-
ax.set_title('Amino Acid Composition Across Datasets')
|
| 373 |
-
ax.set_xlabel('Amino Acid')
|
| 374 |
-
ax.set_ylabel('Relative Frequency')
|
|
|
|
|
|
|
|
|
|
| 375 |
|
| 376 |
fig_individual.set_tight_layout(True)
|
| 377 |
fig_individual.savefig(savepath)
|
|
@@ -379,6 +419,7 @@ def visualize_splits_aa_composition(train_sequences=None, val_sequences=None, te
|
|
| 379 |
|
| 380 |
### Outer methods for visualizing splits
|
| 381 |
def visualize_splits(train_clusters=None, val_clusters=None, test_clusters=None, benchmark_cluster_reps=None, train_color='#0072B2',val_color='#009E73',test_color='#E69F00',esm_embeddings_path=None, onehot_embeddings_path=None):
|
|
|
|
| 382 |
colormap = {
|
| 383 |
'train': train_color,
|
| 384 |
'val': val_color,
|
|
@@ -413,6 +454,14 @@ def visualize_train_val_test_splits(train_clusters, val_clusters, test_clusters,
|
|
| 413 |
val_sequences = val_clusters['member seq'].tolist()
|
| 414 |
test_sequences = test_clusters['member seq'].tolist()
|
| 415 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 416 |
# Create a combined figure with 3 rows and 3 columns
|
| 417 |
set_font()
|
| 418 |
fig_combined, axs = plt.subplots(3, 3, figsize=(24, 18))
|
|
@@ -445,8 +494,9 @@ def visualize_train_val_test_splits(train_clusters, val_clusters, test_clusters,
|
|
| 445 |
axs[2, 2].axis('off')
|
| 446 |
|
| 447 |
plt.tight_layout()
|
| 448 |
-
fig_combined.
|
| 449 |
-
|
|
|
|
| 450 |
|
| 451 |
def visualize_train_test_splits(train_clusters, test_clusters, benchmark_cluster_reps=None, colormap=None, esm_embeddings_path=None, onehot_embeddings_path=None):
|
| 452 |
if colormap is None: colormap=default_color_map
|
|
@@ -493,8 +543,8 @@ def visualize_train_test_splits(train_clusters, test_clusters, benchmark_cluste
|
|
| 493 |
colormap=colormap, axes=axs[2, 1])
|
| 494 |
|
| 495 |
plt.tight_layout()
|
| 496 |
-
fig_combined.savefig('splits/combined_plot.png')
|
| 497 |
-
log_update(f"\nSaved combined figure to splits/combined_plot.png")
|
| 498 |
|
| 499 |
def visualize_train_val_splits(train_clusters, val_clusters, benchmark_cluster_reps=None, colormap=None, esm_embeddings_path=None, onehot_embeddings_path=None):
|
| 500 |
if colormap is None: colormap=default_color_map
|
|
@@ -541,5 +591,5 @@ def visualize_train_val_splits(train_clusters, val_clusters, benchmark_cluster_r
|
|
| 541 |
colormap=colormap, axes=axs[2, 1])
|
| 542 |
|
| 543 |
plt.tight_layout()
|
| 544 |
-
fig_combined.savefig('splits/combined_plot.png')
|
| 545 |
-
log_update(f"\nSaved combined figure to splits/combined_plot.png")
|
|
|
|
| 34 |
# Set the font family globally to Ubuntu
|
| 35 |
plt.rcParams['font.family'] = regular_font.get_name()
|
| 36 |
|
| 37 |
+
# Set the font family globally to Ubuntu
|
| 38 |
+
plt.rcParams['font.family'] = regular_font.get_name()
|
| 39 |
plt.rcParams['mathtext.fontset'] = 'custom'
|
| 40 |
plt.rcParams['mathtext.rm'] = regular_font.get_name()
|
| 41 |
+
plt.rcParams['mathtext.it'] = italic_font.get_name()
|
| 42 |
+
plt.rcParams['mathtext.bf'] = bold_font.get_name()
|
| 43 |
|
| 44 |
global default_color_map
|
| 45 |
default_color_map = {
|
|
|
|
| 99 |
counts = [sequence.count(base) for base in bases]
|
| 100 |
return entropy(counts, base=2)
|
| 101 |
|
| 102 |
+
def visualize_splits_hist(train_lengths=None, val_lengths=None, test_lengths=None, colormap=None, savepath=f'splits/split_vis/length_distributions.png', axes=None):
|
| 103 |
"""
|
| 104 |
Works to plot train, val, test; train, val; or train, test
|
| 105 |
"""
|
|
|
|
| 119 |
total_plots-=1
|
| 120 |
|
| 121 |
# Create a figure and axes with 1 row and 3 columns
|
| 122 |
+
fig_individual, axes_individual = plt.subplots(1, total_plots, figsize=(8*total_plots, 8))
|
| 123 |
|
| 124 |
# Set axes list
|
| 125 |
axes_list = [axes_individual] if axes is None else [axes_individual, axes]
|
|
|
|
| 130 |
for cur_axes in axes_list:
|
| 131 |
# Plot the first histogram
|
| 132 |
cur_axes[0].hist(train_lengths, bins=20, edgecolor='k',color=colormap['train'])
|
| 133 |
+
cur_axes[0].set_xlabel(xlabel, fontsize=24)
|
| 134 |
+
cur_axes[0].set_ylabel(ylabel, fontsize=24)
|
| 135 |
+
cur_axes[0].set_title(f'Train Set Length Distribution (n={len(train_lengths)})', fontsize=24)
|
| 136 |
cur_axes[0].grid(True)
|
| 137 |
cur_axes[0].set_axisbelow(True)
|
| 138 |
+
cur_axes[0].tick_params(axis='x', labelsize=24) # Customize x-axis tick label size
|
| 139 |
+
cur_axes[0].tick_params(axis='y', labelsize=24) # Customize y-axis tick label size
|
| 140 |
|
| 141 |
# Plot the second histogram
|
| 142 |
if not(val_plot_index is None):
|
| 143 |
cur_axes[val_plot_index].hist(val_lengths, bins=20, edgecolor='k',color=colormap['val'])
|
| 144 |
+
cur_axes[val_plot_index].set_xlabel(xlabel, fontsize=24)
|
| 145 |
+
cur_axes[val_plot_index].set_ylabel(ylabel, fontsize=24)
|
| 146 |
+
cur_axes[val_plot_index].set_title(f'Validation Set Length Distribution (n={len(val_lengths)})', fontsize=24)
|
| 147 |
cur_axes[val_plot_index].grid(True)
|
| 148 |
cur_axes[val_plot_index].set_axisbelow(True)
|
| 149 |
+
cur_axes[val_plot_index].tick_params(axis='x', labelsize=24)
|
| 150 |
+
cur_axes[val_plot_index].tick_params(axis='y', labelsize=24)
|
| 151 |
|
| 152 |
# Plot the third histogram
|
| 153 |
if not(test_plot_index is None):
|
| 154 |
cur_axes[test_plot_index].hist(test_lengths, bins=20, edgecolor='k',color=colormap['test'])
|
| 155 |
+
cur_axes[test_plot_index].set_xlabel(xlabel, fontsize=24)
|
| 156 |
+
cur_axes[test_plot_index].set_ylabel(ylabel, fontsize=24)
|
| 157 |
+
cur_axes[test_plot_index].set_title(f'Test Set Length Distribution (n={len(test_lengths)})', fontsize=24)
|
| 158 |
cur_axes[test_plot_index].grid(True)
|
| 159 |
cur_axes[test_plot_index].set_axisbelow(True)
|
| 160 |
+
cur_axes[test_plot_index].tick_params(axis='x', labelsize=24)
|
| 161 |
+
cur_axes[test_plot_index].tick_params(axis='y', labelsize=24)
|
| 162 |
|
| 163 |
# Adjust layout
|
| 164 |
fig_individual.set_tight_layout(True)
|
|
|
|
| 167 |
fig_individual.savefig(savepath)
|
| 168 |
log_update(f"\tSaved figure to {savepath}")
|
| 169 |
|
| 170 |
+
def visualize_splits_scatter(train_clusters=None, val_clusters=None, test_clusters=None, benchmark_cluster_reps=None, colormap=None, savepath='splits/split_vis/scatterplot.png', axes=None):
|
| 171 |
set_font()
|
| 172 |
if colormap is None: colormap=default_color_map
|
| 173 |
|
|
|
|
| 216 |
# Specially mark the benchmark clusters because these can't be reallocated
|
| 217 |
for ax in axes_list:
|
| 218 |
ax.plot(train_clustersgb['cluster size (n_members)'],train_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['train'],label='train')
|
| 219 |
+
train_clustersgb.to_csv(savepath.replace(".png","_train_source_data.csv"),index=False)
|
| 220 |
if not(val_clusters is None):
|
| 221 |
ax.plot(val_clustersgb['cluster size (n_members)'],val_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['val'],label='val')
|
| 222 |
+
val_clustersgb.to_csv(savepath.replace(".png","_val_source_data.csv"),index=False)
|
| 223 |
if not(test_clusters is None):
|
| 224 |
ax.plot(test_clustersgb['cluster size (n_members)'],test_clustersgb['percent_proteins'],linestyle='None',marker='.',color=colormap['test'],label='test')
|
| 225 |
+
test_clustersgb.to_csv(savepath.replace(".png","_test_source_data.csv"),index=False)
|
| 226 |
if not(benchmark_cluster_reps is None):
|
| 227 |
ax.plot(benchmark_clustersgb['cluster size (n_members)'],benchmark_clustersgb['percent_proteins'],
|
| 228 |
marker='o',
|
|
|
|
| 232 |
markeredgewidth=1.5,
|
| 233 |
label='benchmark'
|
| 234 |
)
|
| 235 |
+
benchmark_clustersgb.to_csv(savepath.replace(".png","_benchmark_source_data.csv"),index=False)
|
| 236 |
+
ax.set_ylabel('Percentage of Proteins in Dataset', fontsize=24)
|
| 237 |
+
ax.set_xlabel('Cluster Size', fontsize=24)
|
| 238 |
+
ax.tick_params(axis='x', labelsize=24) # Customize x-axis tick label size
|
| 239 |
+
ax.tick_params(axis='y', labelsize=24) # Customize y-axis tick label size
|
| 240 |
+
|
| 241 |
+
ax.legend(fontsize=24,markerscale=4)
|
| 242 |
|
| 243 |
# save the figure
|
| 244 |
fig_individual.set_tight_layout(True)
|
|
|
|
| 246 |
log_update(f"\tSaved figure to {savepath}")
|
| 247 |
|
| 248 |
|
| 249 |
+
def visualize_splits_tsne(train_sequences=None, val_sequences=None, test_sequences=None, colormap=None, esm_type="esm2_t33_650M_UR50D", embedding_path="fuson_db_embeddings/fuson_db_esm2_t33_650M_UR50D_avg_embeddings.pkl", savepath='splits/split_vis/tsne_plot.png',axes=None):
|
| 250 |
set_font()
|
| 251 |
|
| 252 |
if colormap is None: colormap=default_color_map
|
|
|
|
| 300 |
fig_individual.savefig(savepath)
|
| 301 |
log_update(f"\tSaved figure to {savepath}")
|
| 302 |
|
| 303 |
+
def visualize_splits_shannon_entropy(train_sequences=None, val_sequences=None, test_sequences=None, colormap=None, savepath='splits/split_vis/shannon_entropy_plot.png',axes=None):
|
| 304 |
set_font()
|
| 305 |
"""
|
| 306 |
Generate Shannon entropy plots for train, validation, and test sets.
|
|
|
|
| 331 |
|
| 332 |
for ax in axes_list:
|
| 333 |
ax[0].hist(train_entropy, bins=20, edgecolor='k', color=colormap['train'])
|
| 334 |
+
ax[0].set_title(f'Train Set (n={len(train_entropy)})', fontsize=24)
|
| 335 |
+
ax[0].set_xlabel('Shannon Entropy', fontsize=24)
|
| 336 |
+
ax[0].set_ylabel('Frequency', fontsize=24)
|
| 337 |
ax[0].grid(True)
|
| 338 |
ax[0].set_axisbelow(True)
|
| 339 |
+
axes[0].tick_params(axis='x', labelsize=24)
|
| 340 |
+
axes[0].tick_params(axis='y', labelsize=24)
|
| 341 |
+
|
| 342 |
+
train_shannon_source_data = pd.DataFrame(data={
|
| 343 |
+
'sequence': train_sequences, 'shannon_entropy': train_entropy
|
| 344 |
+
})
|
| 345 |
+
train_shannon_source_data.to_csv(savepath.replace(".png","_train_source_data.csv"),index=False)
|
| 346 |
|
| 347 |
if not(val_plot_index is None):
|
| 348 |
ax[val_plot_index].hist(val_entropy, bins=20, edgecolor='k', color=colormap['val'])
|
| 349 |
+
ax[val_plot_index].set_title(f'Validation Set (n={len(val_entropy)})', fontsize=24)
|
| 350 |
+
ax[val_plot_index].set_xlabel('Shannon Entropy', fontsize=24)
|
| 351 |
ax[val_plot_index].grid(True)
|
| 352 |
ax[val_plot_index].set_axisbelow(True)
|
| 353 |
+
ax[val_plot_index].tick_params(axis='x', labelsize=24)
|
| 354 |
+
ax[val_plot_index].tick_params(axis='y', labelsize=24)
|
| 355 |
+
|
| 356 |
+
val_shannon_source_data = pd.DataFrame(data={
|
| 357 |
+
'sequence': val_sequences, 'shannon_entropy': val_entropy
|
| 358 |
+
})
|
| 359 |
+
val_shannon_source_data.to_csv(savepath.replace(".png","_val_source_data.csv"),index=False)
|
| 360 |
+
|
| 361 |
if not(test_plot_index is None):
|
| 362 |
ax[test_plot_index].hist(test_entropy, bins=20, edgecolor='k', color=colormap['test'])
|
| 363 |
+
ax[test_plot_index].set_title(f'Test Set (n={len(test_entropy)})', fontsize=24)
|
| 364 |
+
ax[test_plot_index].set_xlabel('Shannon Entropy', fontsize=24)
|
| 365 |
ax[test_plot_index].grid(True)
|
| 366 |
ax[test_plot_index].set_axisbelow(True)
|
| 367 |
+
ax[test_plot_index].tick_params(axis='x', labelsize=24)
|
| 368 |
+
ax[test_plot_index].tick_params(axis='y', labelsize=24)
|
| 369 |
+
|
| 370 |
+
test_shannon_source_data = pd.DataFrame(data={
|
| 371 |
+
'sequence': test_sequences, 'shannon_entropy': test_entropy
|
| 372 |
+
})
|
| 373 |
+
test_shannon_source_data.to_csv(savepath.replace(".png","_test_source_data.csv"),index=False)
|
| 374 |
|
| 375 |
fig_individual.set_tight_layout(True)
|
| 376 |
fig_individual.savefig(savepath)
|
| 377 |
log_update(f"\tSaved figure to {savepath}")
|
| 378 |
|
| 379 |
+
def visualize_splits_aa_composition(train_sequences=None, val_sequences=None, test_sequences=None, colormap=None, savepath='splits/split_vis/aa_comp.png',axes=None):
|
| 380 |
set_font()
|
| 381 |
if colormap is None: colormap=default_color_map
|
| 382 |
|
|
|
|
| 401 |
if (val_sequences is None) and not(test_sequences is None):
|
| 402 |
comp_df = pd.DataFrame([train_comp, test_comp], index=['train', 'test']).T
|
| 403 |
colors = [colormap[col] for col in comp_df.columns]
|
| 404 |
+
comp_df.to_csv(savepath.replace(".png","_source_data.csv"))
|
| 405 |
|
| 406 |
# Plotting
|
| 407 |
for ax in axes_list:
|
| 408 |
comp_df.plot(kind='bar', color=colors, ax=ax)
|
| 409 |
+
ax.set_title('Amino Acid Composition Across Datasets', fontsize=24)
|
| 410 |
+
ax.set_xlabel('Amino Acid', fontsize=24)
|
| 411 |
+
ax.set_ylabel('Relative Frequency', fontsize=24)
|
| 412 |
+
ax.tick_params(axis='x', labelsize=24) # Customize x-axis tick label size
|
| 413 |
+
ax.tick_params(axis='y', labelsize=24) # Customize y-axis tick label size
|
| 414 |
+
ax.legend(fontsize=16, markerscale=2)
|
| 415 |
|
| 416 |
fig_individual.set_tight_layout(True)
|
| 417 |
fig_individual.savefig(savepath)
|
|
|
|
| 419 |
|
| 420 |
### Outer methods for visualizing splits
|
| 421 |
def visualize_splits(train_clusters=None, val_clusters=None, test_clusters=None, benchmark_cluster_reps=None, train_color='#0072B2',val_color='#009E73',test_color='#E69F00',esm_embeddings_path=None, onehot_embeddings_path=None):
|
| 422 |
+
os.makedirs("splits/split_vis",exist_ok=True)
|
| 423 |
colormap = {
|
| 424 |
'train': train_color,
|
| 425 |
'val': val_color,
|
|
|
|
| 454 |
val_sequences = val_clusters['member seq'].tolist()
|
| 455 |
test_sequences = test_clusters['member seq'].tolist()
|
| 456 |
|
| 457 |
+
# save length source data
|
| 458 |
+
train_lengths_source_data=pd.DataFrame(data={"sequence":train_sequences,"length":train_lengths})
|
| 459 |
+
train_lengths_source_data.to_csv("splits/split_vis/train_lengths_source_data.csv",index=False)
|
| 460 |
+
val_lengths_source_data=pd.DataFrame(data={"sequence":val_sequences,"length":val_lengths})
|
| 461 |
+
val_lengths_source_data.to_csv("splits/split_vis/val_lengths_source_data.csv",index=False)
|
| 462 |
+
test_lengths_source_data=pd.DataFrame(data={"sequence":test_sequences,"length":test_lengths})
|
| 463 |
+
test_lengths_source_data.to_csv("splits/split_vis/test_lengths_source_data.csv",index=False)
|
| 464 |
+
|
| 465 |
# Create a combined figure with 3 rows and 3 columns
|
| 466 |
set_font()
|
| 467 |
fig_combined, axs = plt.subplots(3, 3, figsize=(24, 18))
|
|
|
|
| 494 |
axs[2, 2].axis('off')
|
| 495 |
|
| 496 |
plt.tight_layout()
|
| 497 |
+
fig_combined.set_tight_layout(True)
|
| 498 |
+
fig_combined.savefig('splits/split_vis/combined_plot.png', bbox_inches="tight")
|
| 499 |
+
log_update(f"\nSaved combined figure to splits/split_vis/combined_plot.png")
|
| 500 |
|
| 501 |
def visualize_train_test_splits(train_clusters, test_clusters, benchmark_cluster_reps=None, colormap=None, esm_embeddings_path=None, onehot_embeddings_path=None):
|
| 502 |
if colormap is None: colormap=default_color_map
|
|
|
|
| 543 |
colormap=colormap, axes=axs[2, 1])
|
| 544 |
|
| 545 |
plt.tight_layout()
|
| 546 |
+
fig_combined.savefig('splits/split_vis/combined_plot.png')
|
| 547 |
+
log_update(f"\nSaved combined figure to splits/split_vis/combined_plot.png")
|
| 548 |
|
| 549 |
def visualize_train_val_splits(train_clusters, val_clusters, benchmark_cluster_reps=None, colormap=None, esm_embeddings_path=None, onehot_embeddings_path=None):
|
| 550 |
if colormap is None: colormap=default_color_map
|
|
|
|
| 591 |
colormap=colormap, axes=axs[2, 1])
|
| 592 |
|
| 593 |
plt.tight_layout()
|
| 594 |
+
fig_combined.savefig('splits/split_vis/combined_plot.png')
|
| 595 |
+
log_update(f"\nSaved combined figure to splits/split_vis/combined_plot.png")
|